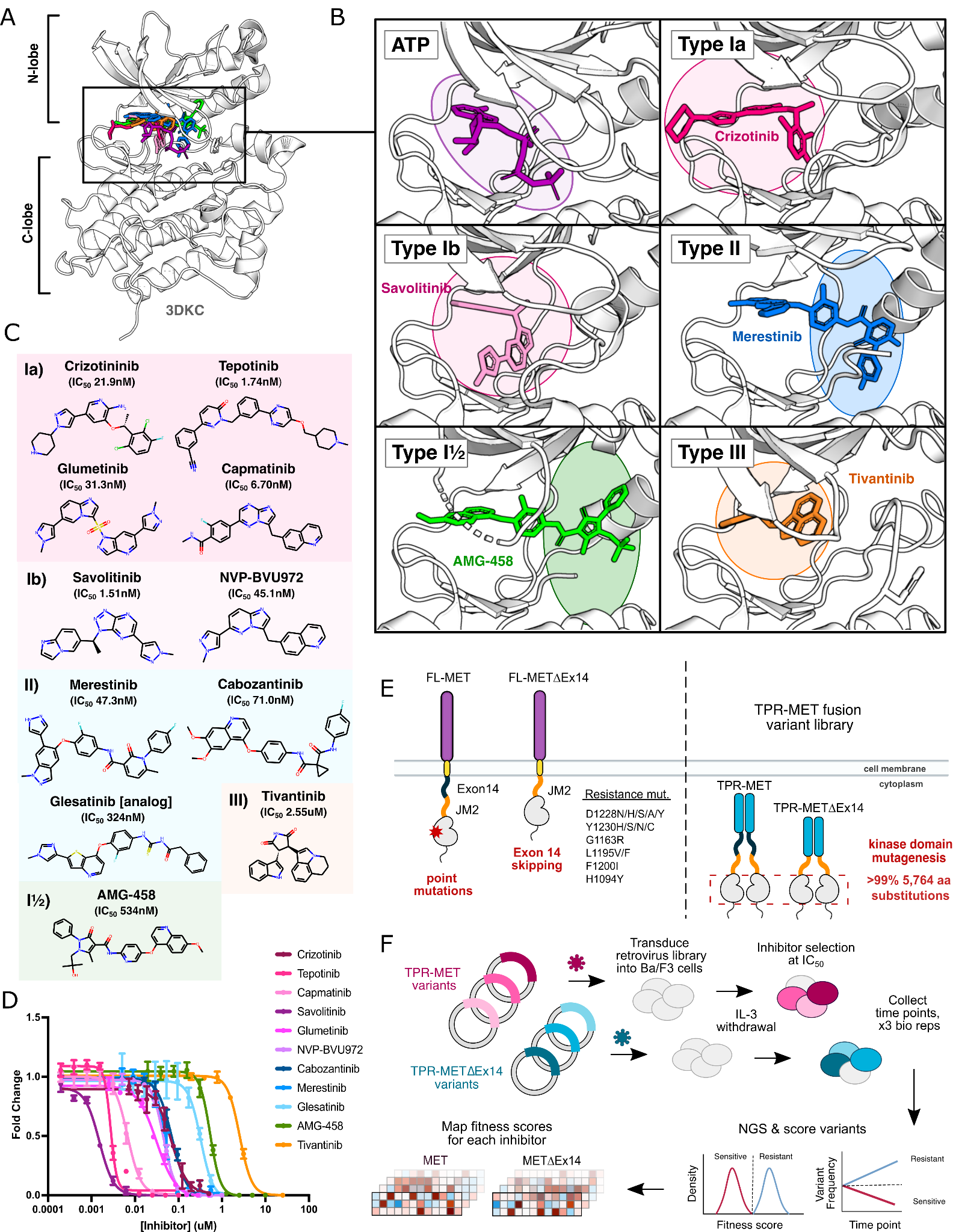
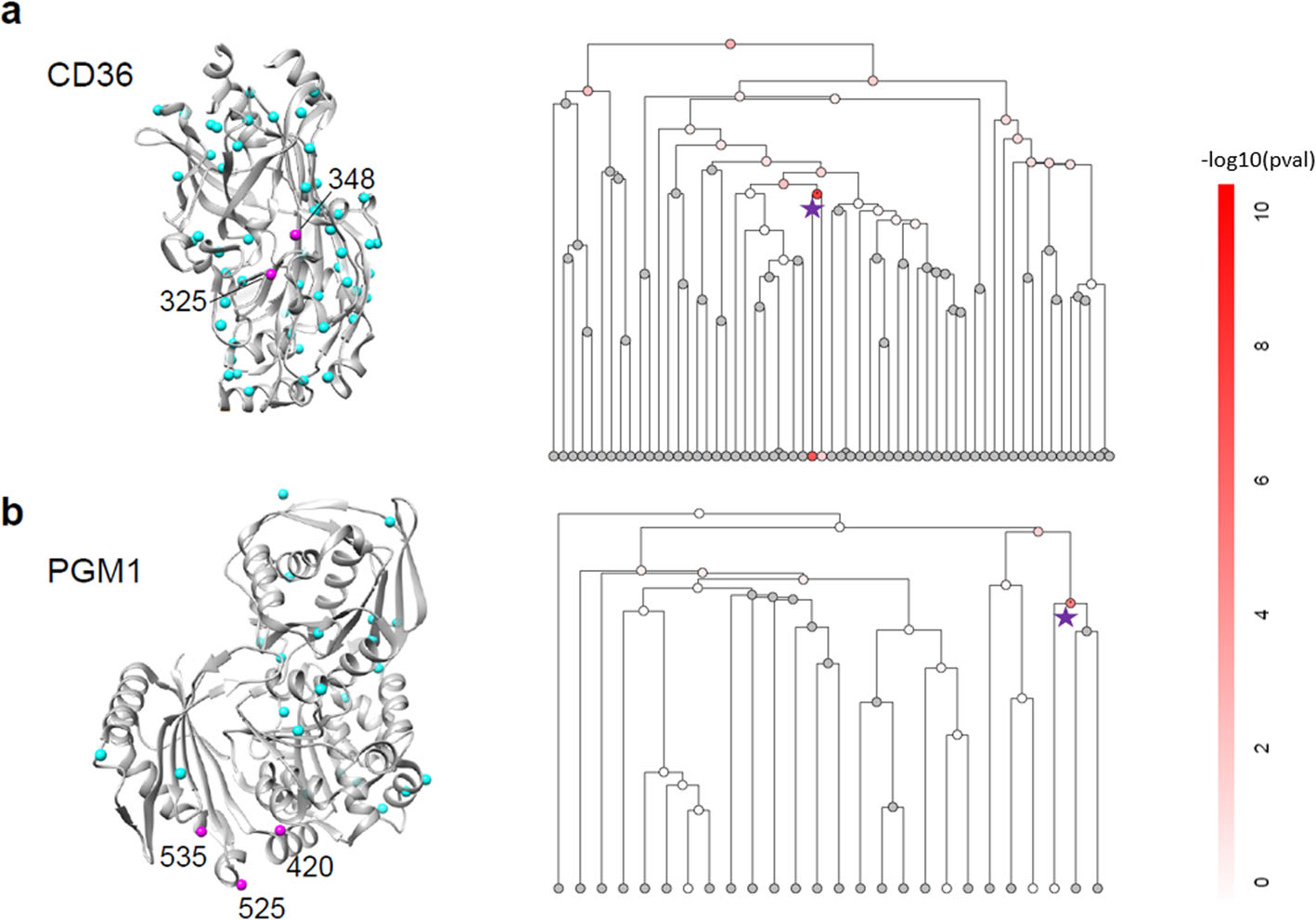
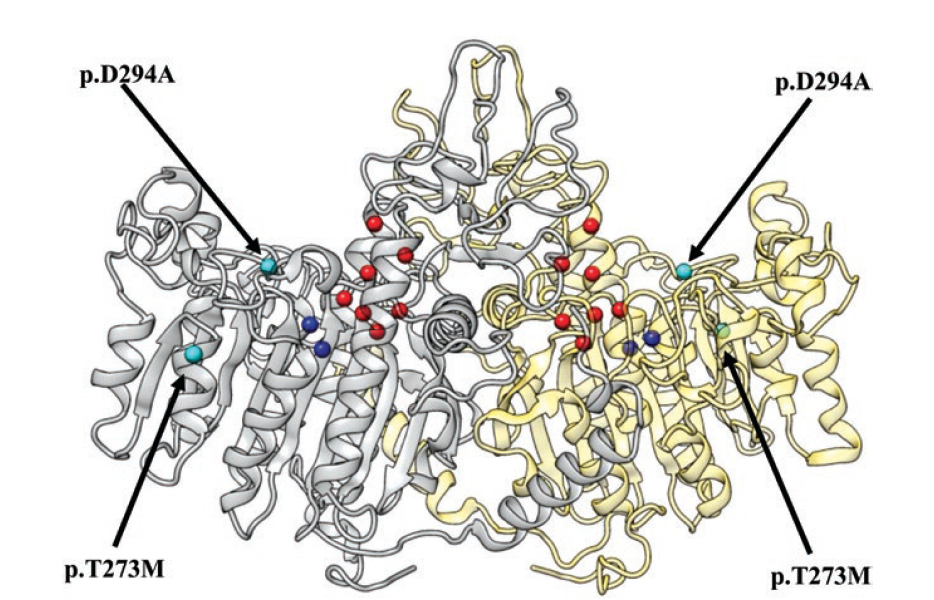
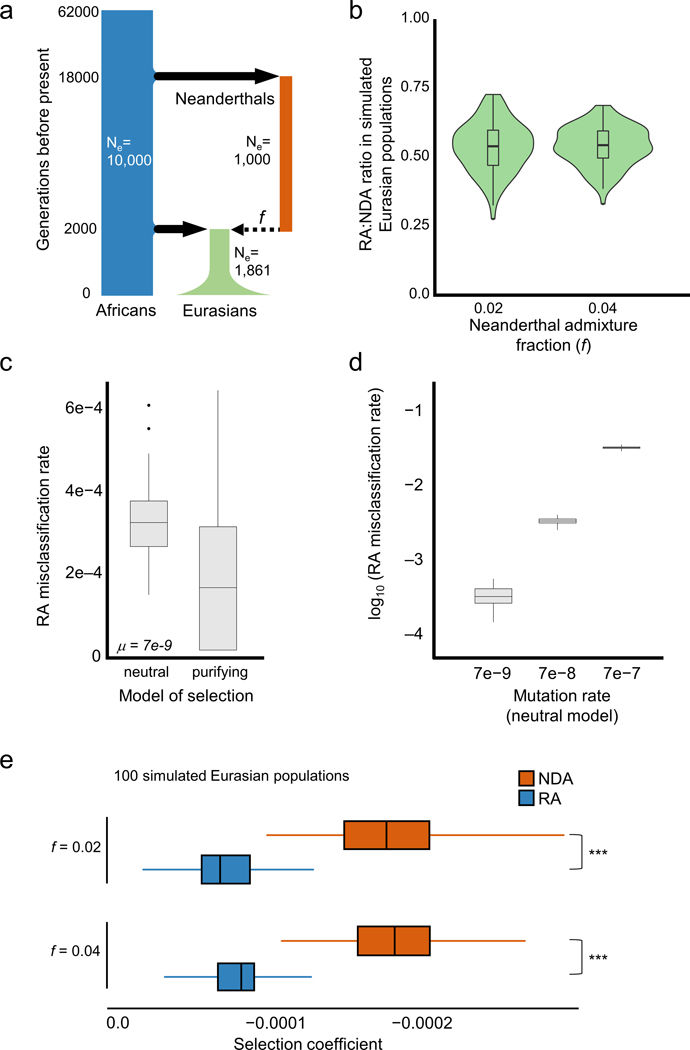
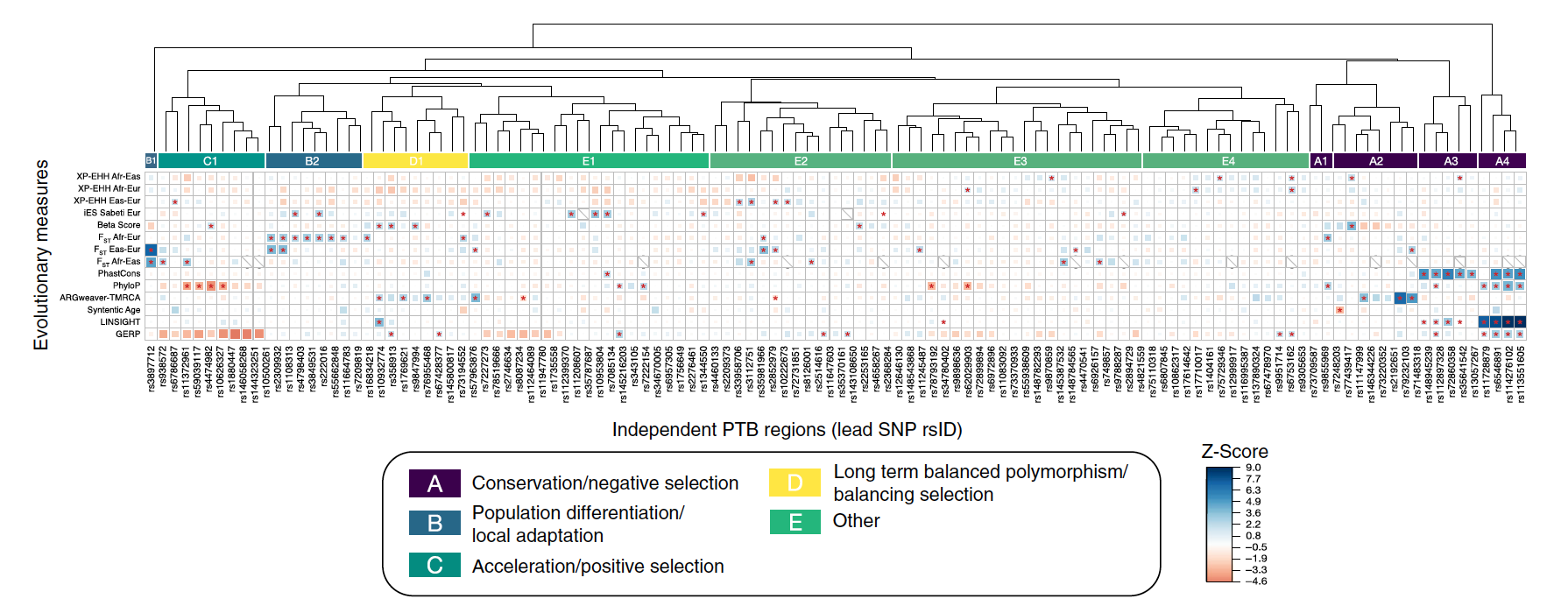
Mapping kinase domain resistance mechanisms for the MET receptor tyrosine kinase via deep mutational scanning
Estevam GO, Linossi EM, Rao J, Macdonald CB, Ravikumar A, Chrispens KM, Capra JA, Coyote-Maestas W, Pimentel H, Collisson EA, Jura N, Fraser JS.*
bioRxiv,
2024
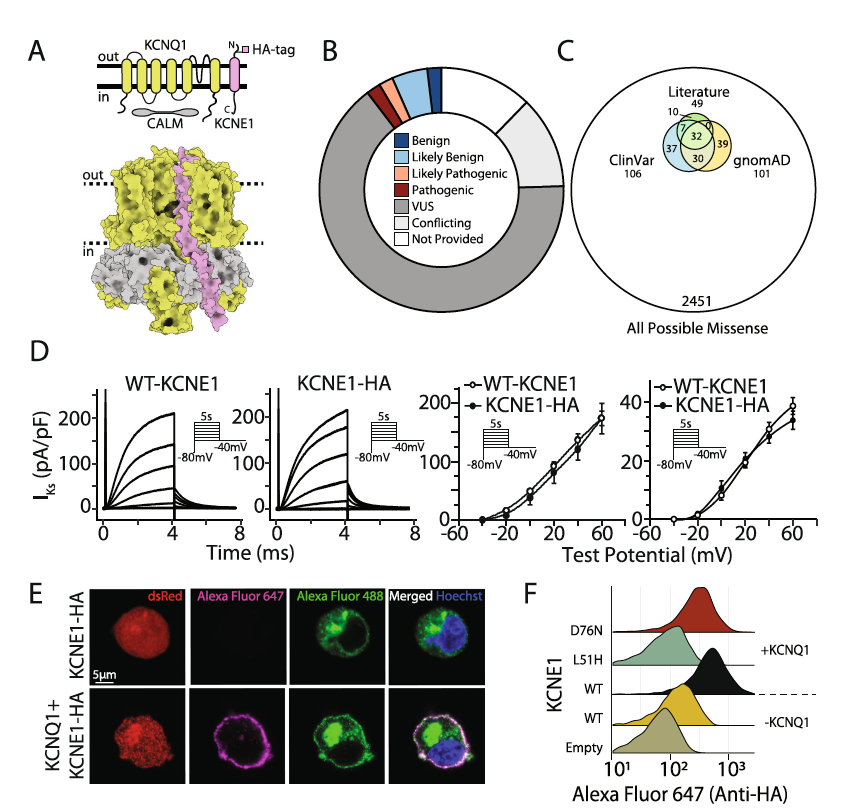
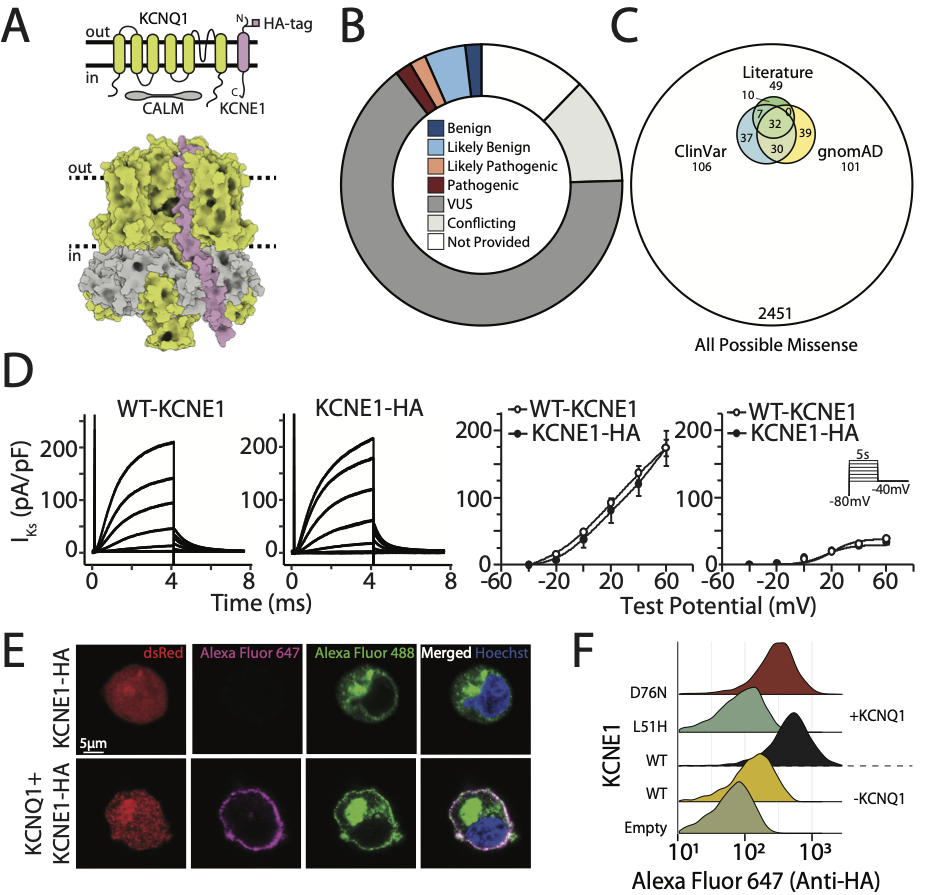
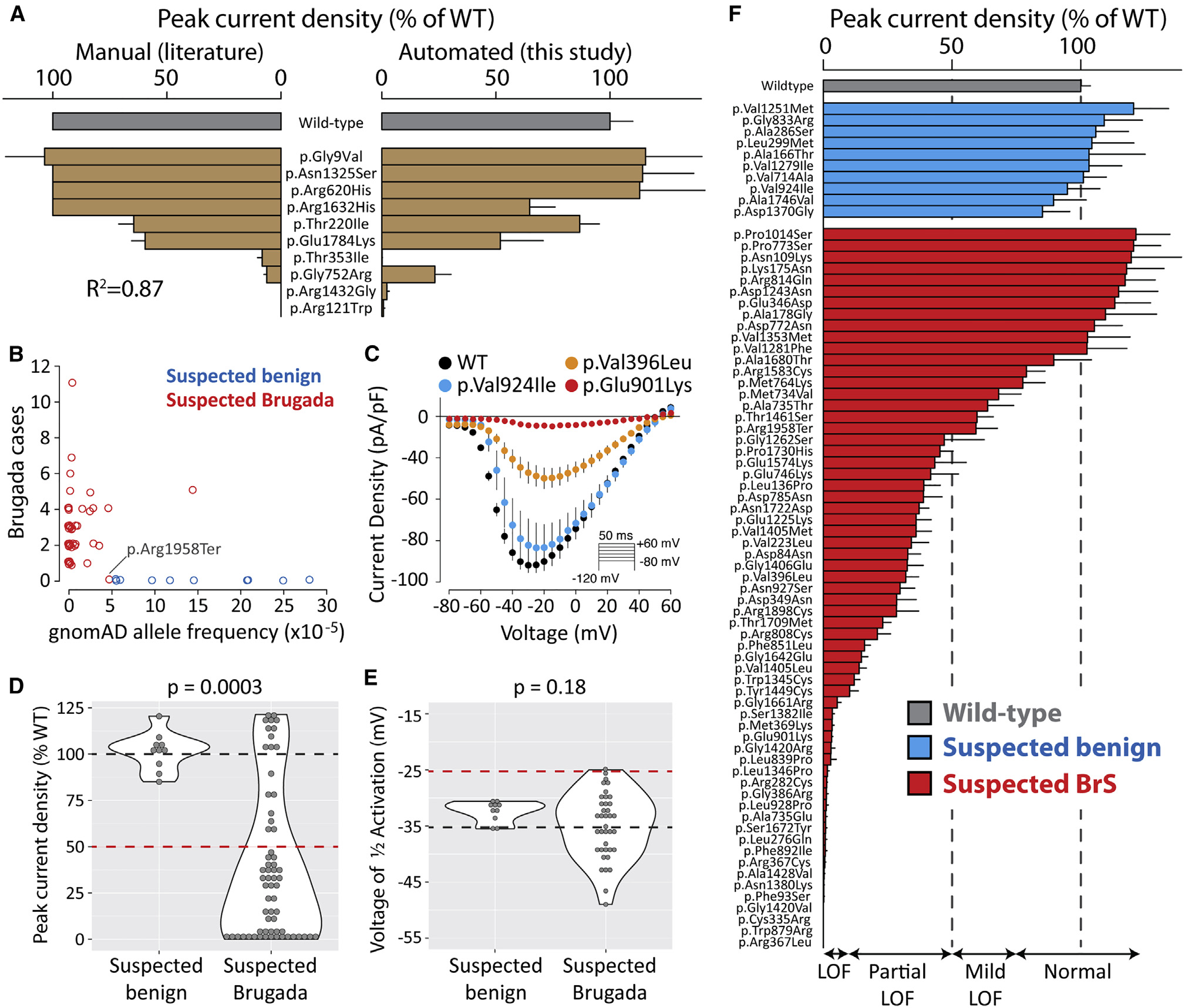
High-throughput functional mapping of variants in an arrhythmia gene, KCNE1, reveals novel biology
Muhammad A, Calandranis ME, Li B, Yang T, Blackwell DJ, Harvey ML, Smith JE, Daniel ZA, Chew AE, Capra JA, Matreyek KA, Fowler DM, Roden DM^, Glazer AM.^*
Genome Med,
2024
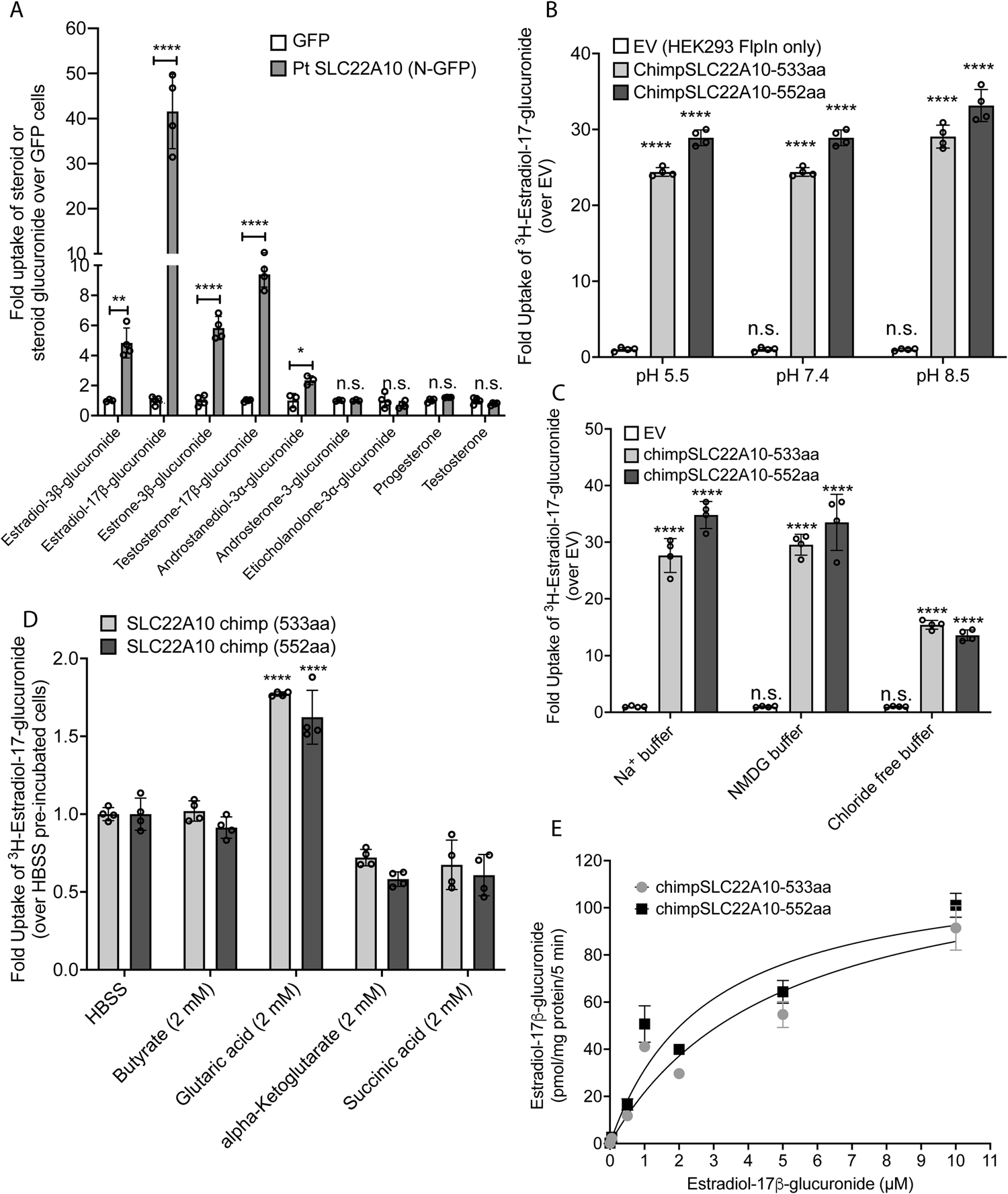
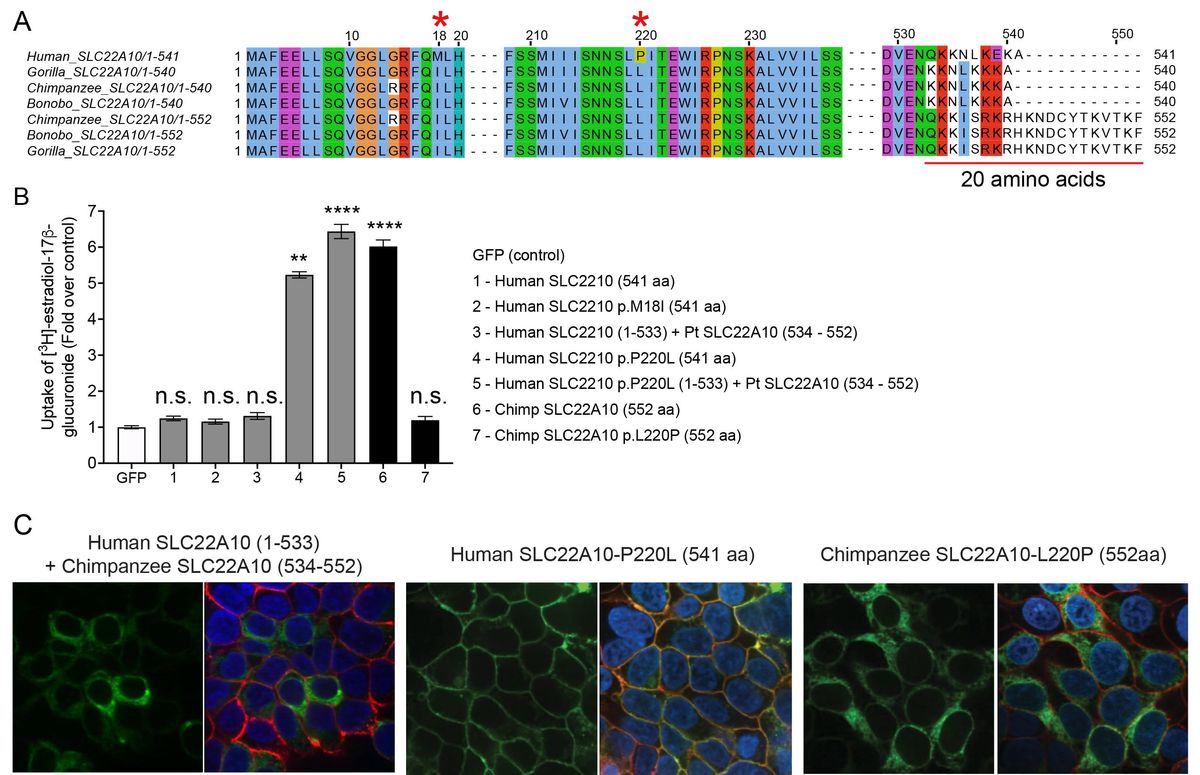
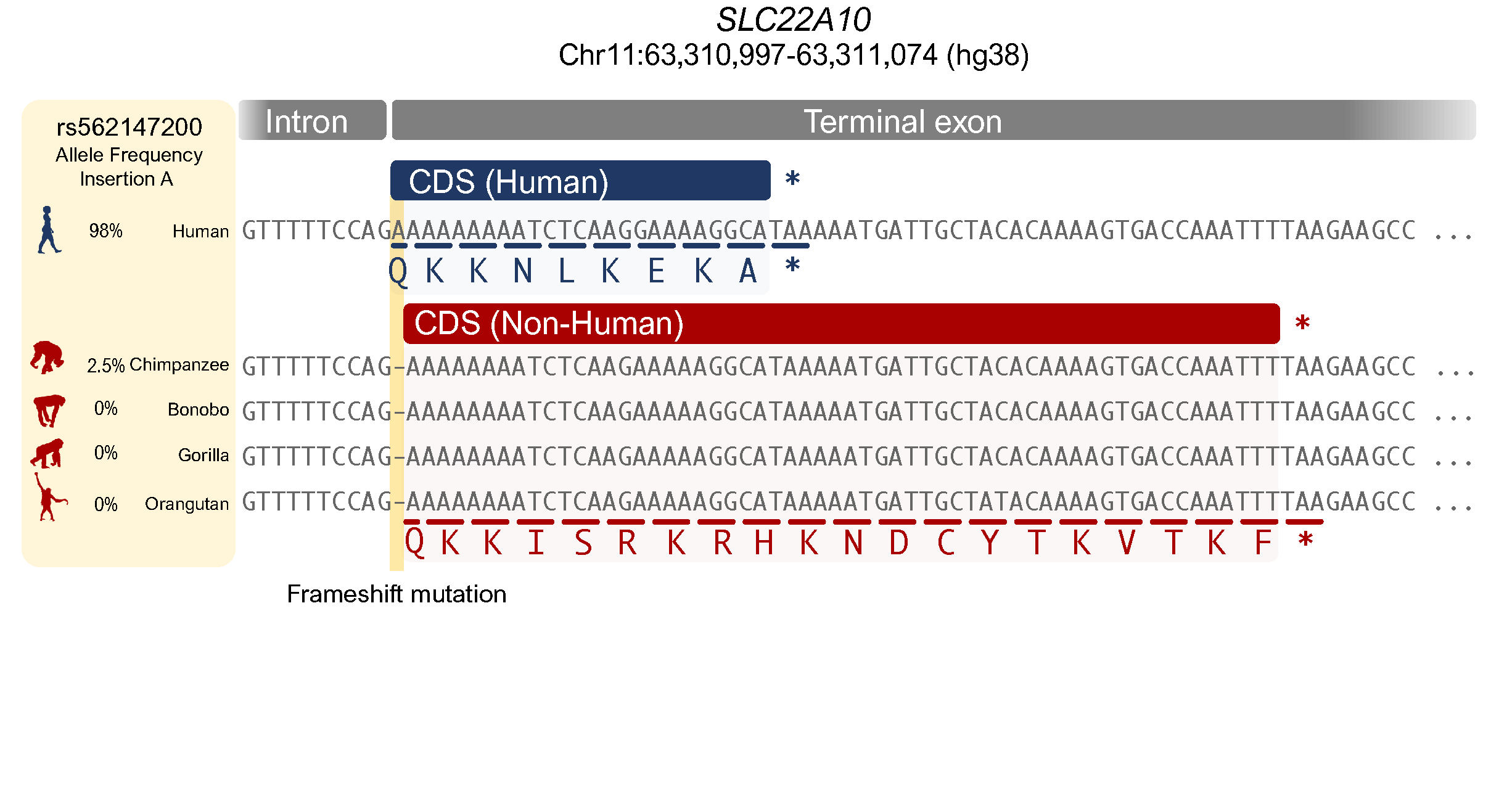
Illuminating the function of the orphan transporter, SLC22A10, in humans and other primates
Yee SW, Ferrández-Peral L, Alentorn-Moron P, Fontsere C, Ceylan M, Koleske ML, Handin N, Artegoitia VM, Lara G, Chien HC, Zhou X, Dainat J, Zalevsky A, Sali A, Brand CM, Wolfreys FD, Yang J, Gestwicki JE, Capra JA, Artursson P, Newman JW, Marquès-Bonet T, Giacomini KM.*
Nat Commun,
2024
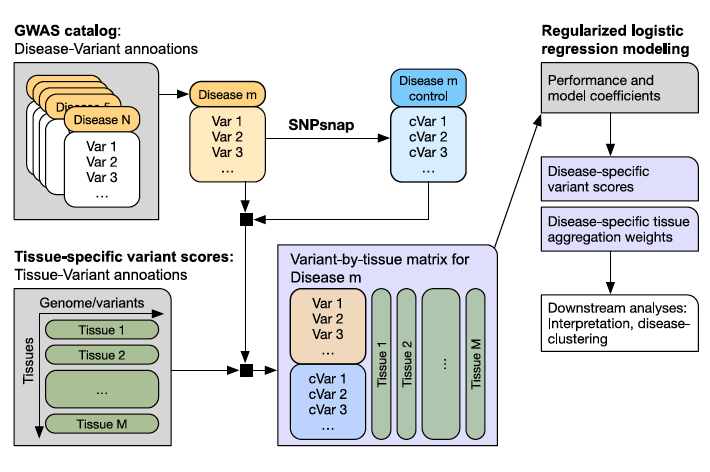
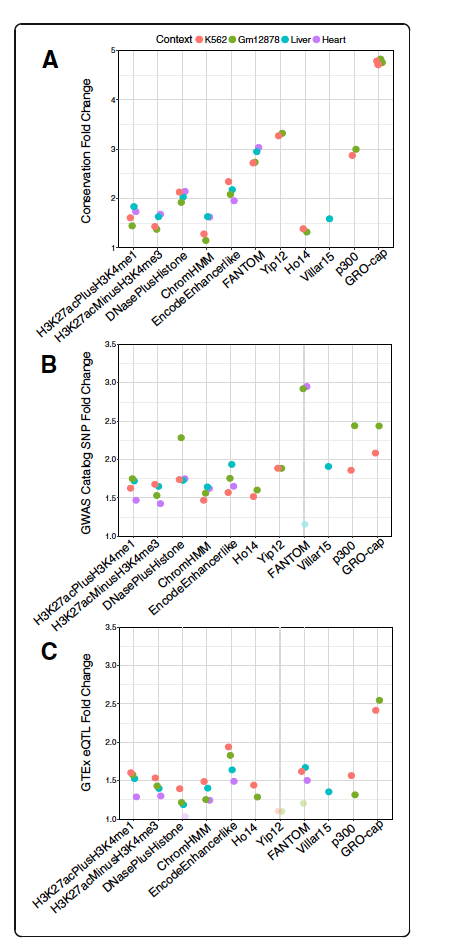
Disease-specific prioritization of non-coding GWAS variants based on chromatin accessibility
Liang Q, Abraham A, Capra JA, Kostka D.*
HGG Adv,
2024
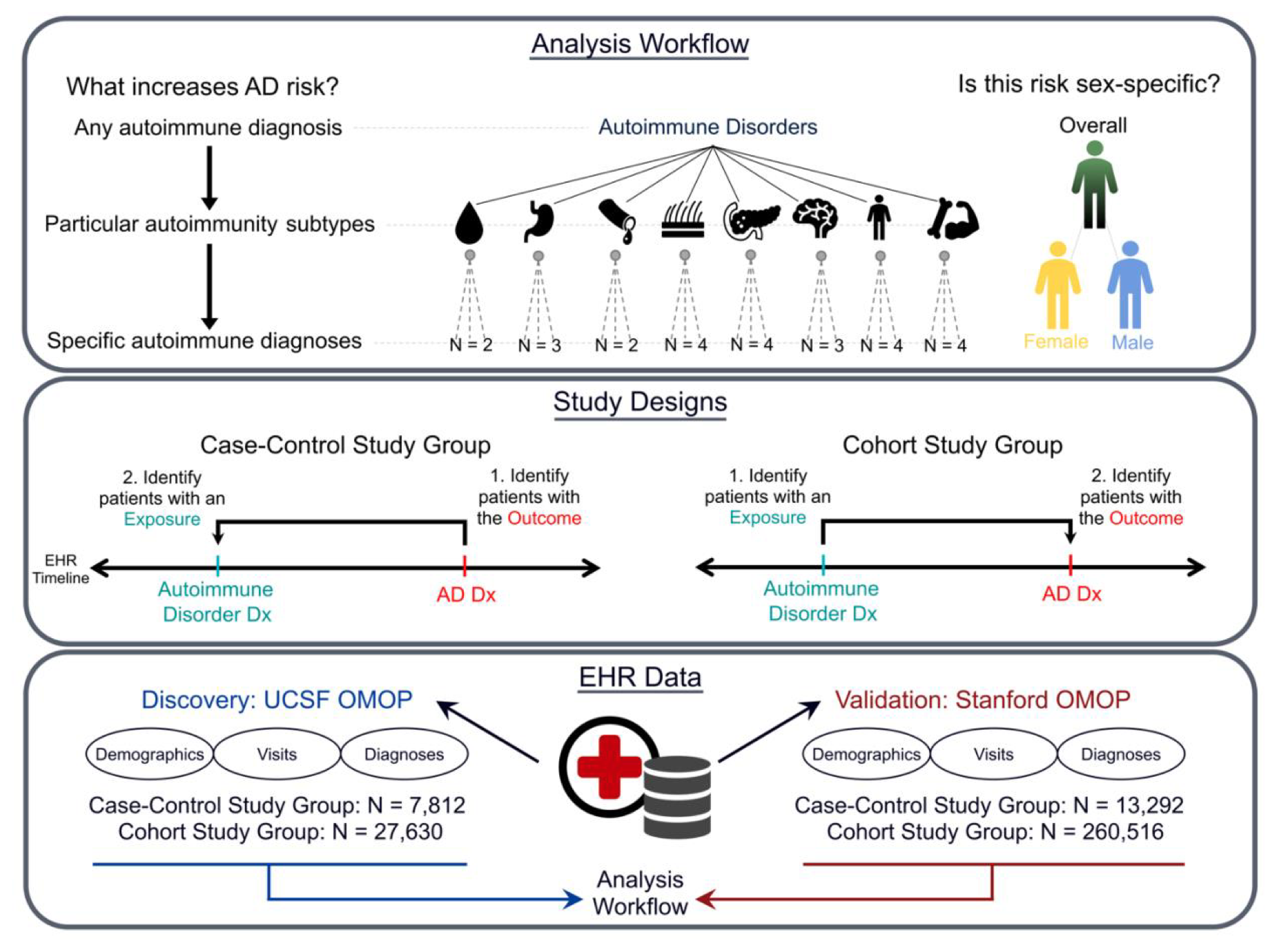
Exposure to autoimmune disorders increases Alzheimer’s disease risk in a multi-site electronic health record analysis
Ramey GD, Tang A, Phongpreecha T, Yang MM, Woldemariam SR, Oskotsky TT, Montine TJ, Allen I, Miller ZA, Aghaeepour N, Capra JA*, Sirota M.*
medRxiv,
2024
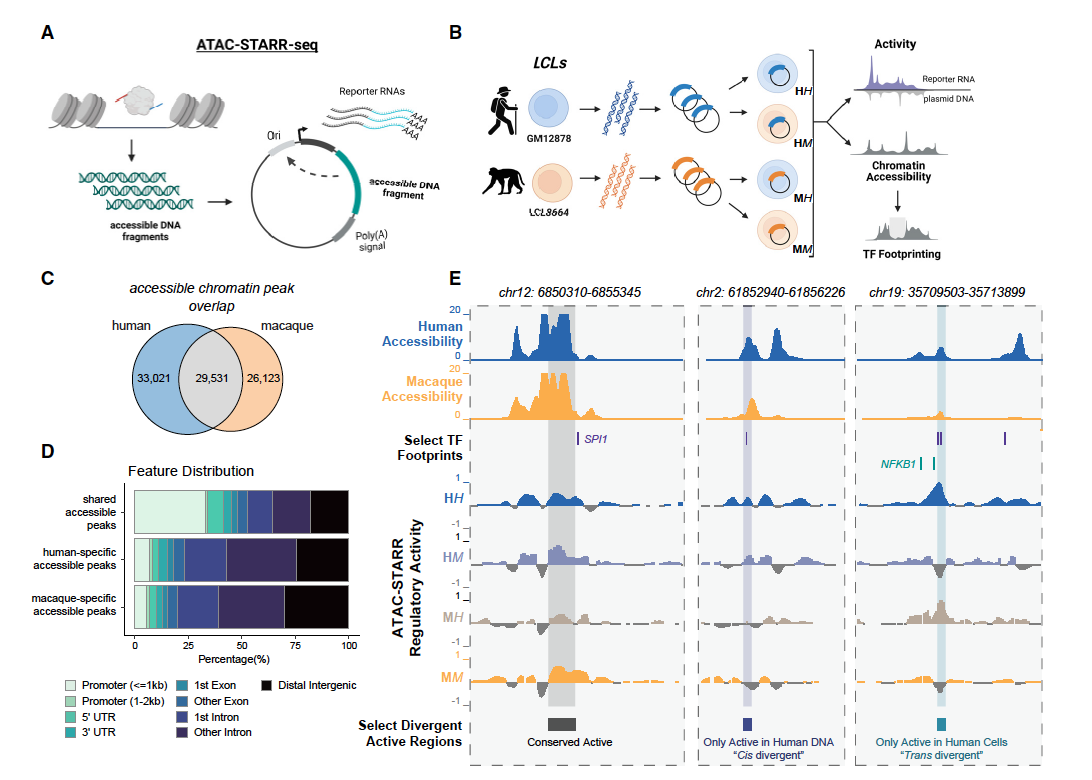
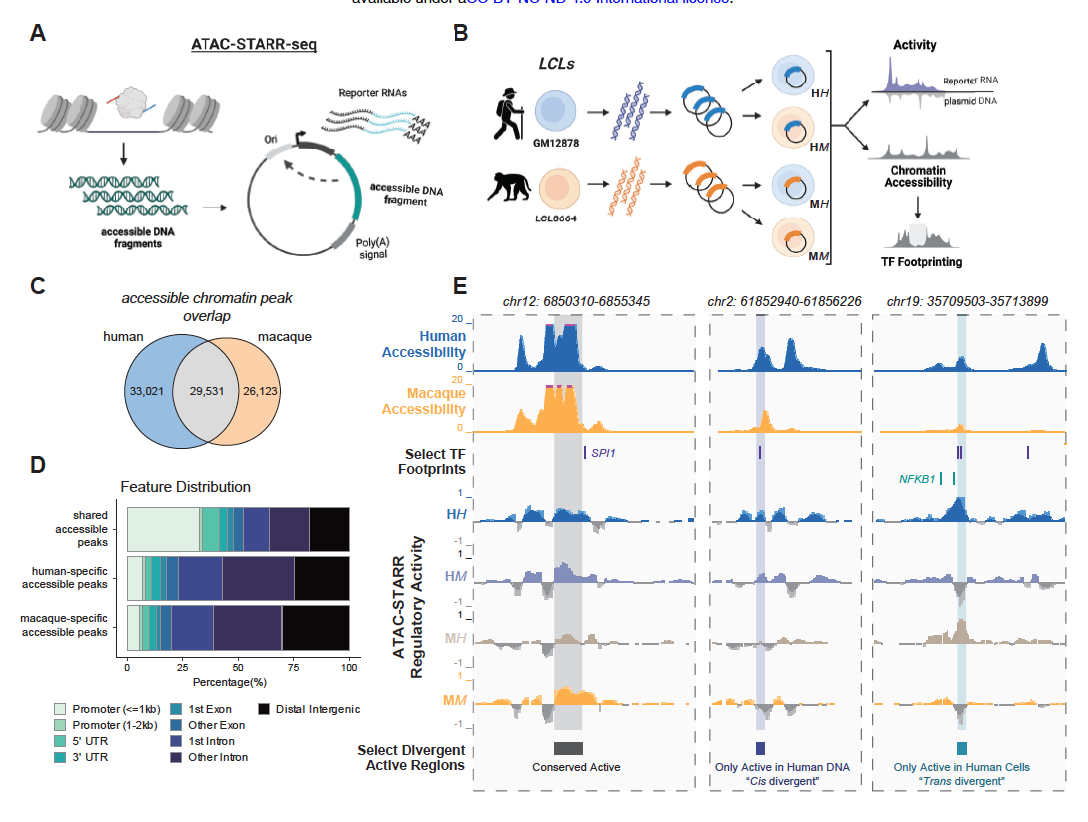
Human gene regulatory evolution is driven by the divergence of regulatory element function in both cis and trans
Hansen TJ, Fong SL, Day JK, Capra JA*, Hodges E.*
Cell Genom,
2024
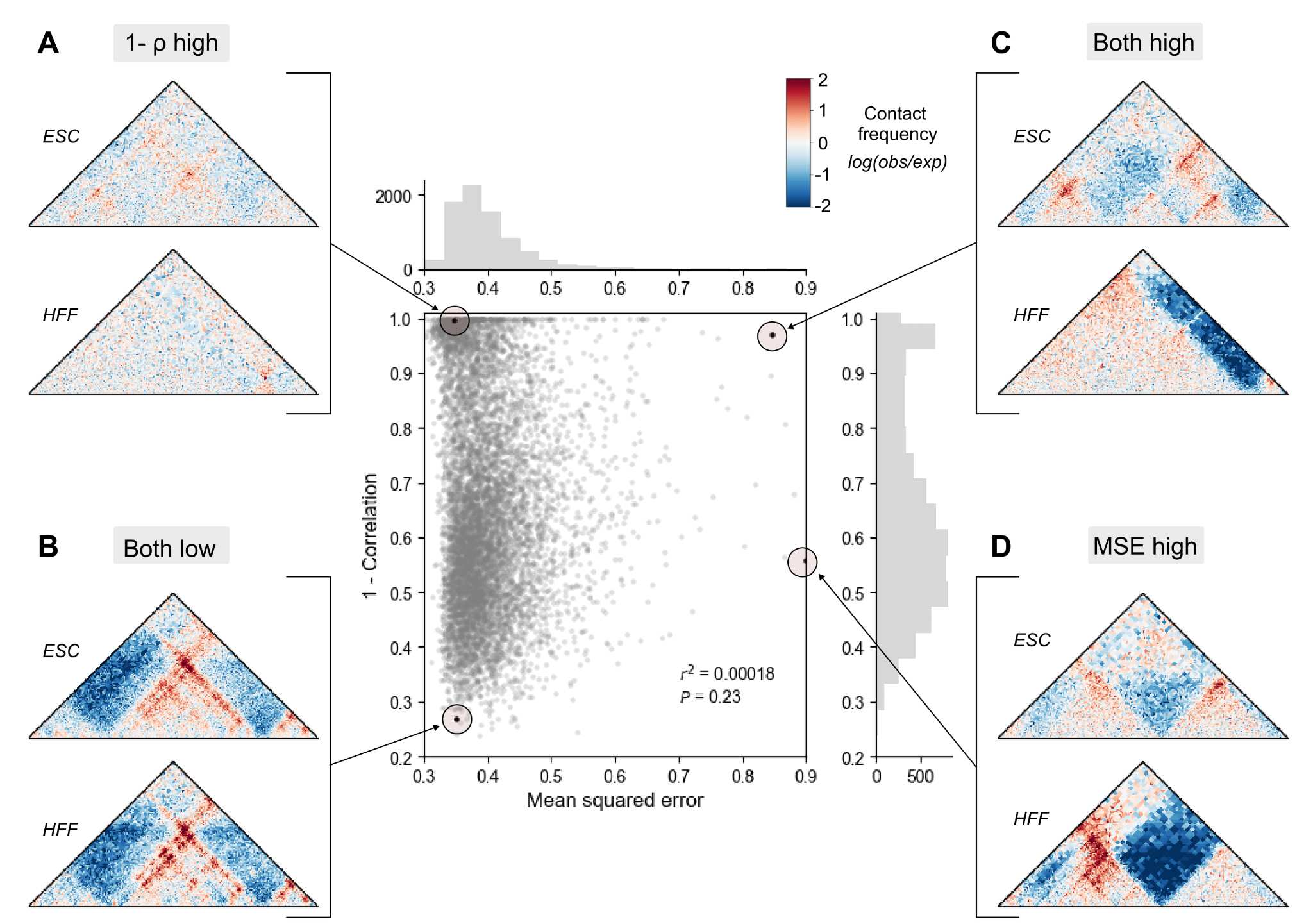
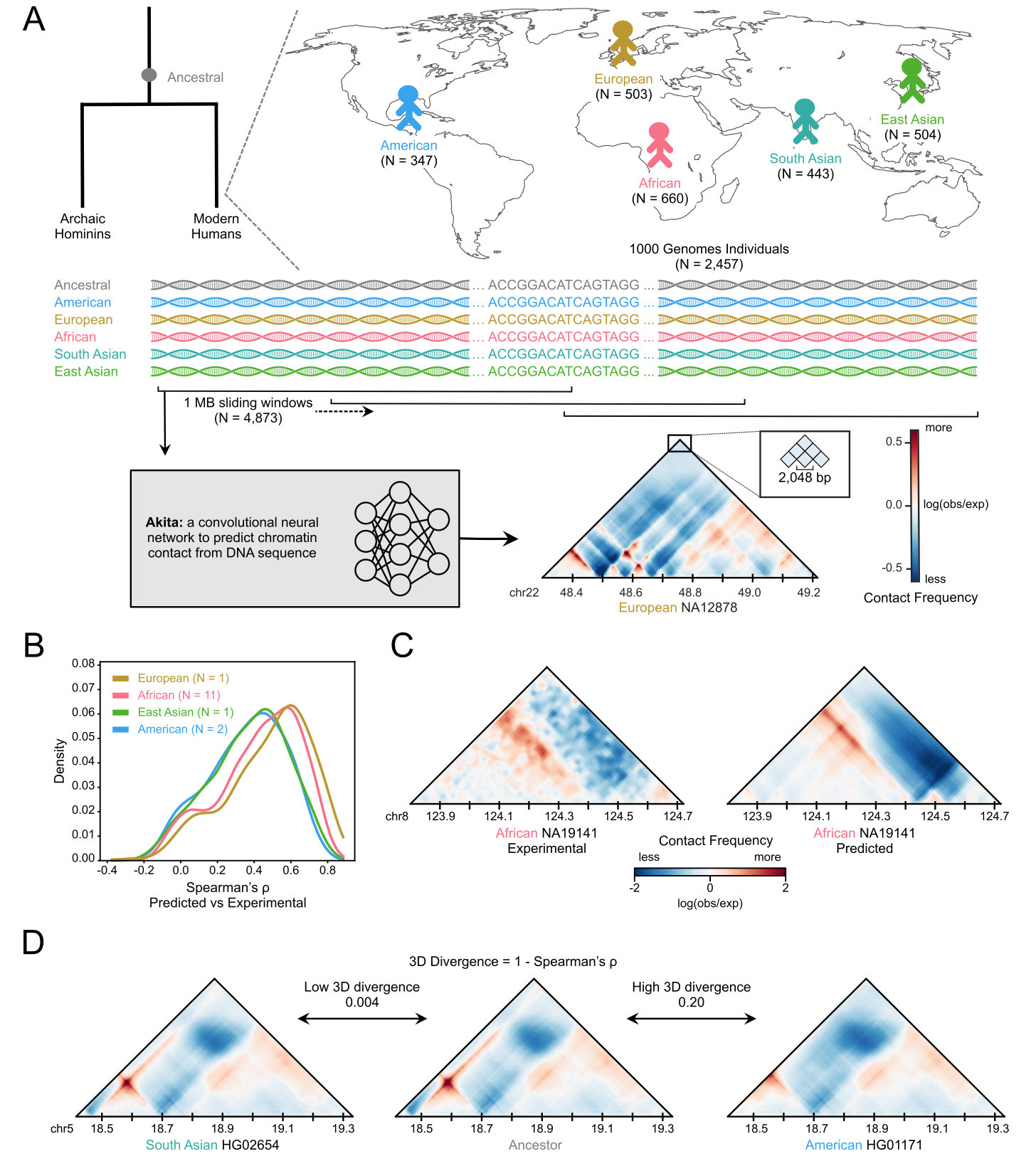
Machine learning reveals the diversity of human 3D chromatin contact patterns
Gilbertson EN, Brand CM, McArthur E, Rinker DC, Kuang S, Pollard KS, Capra JA*
bioRxiv,
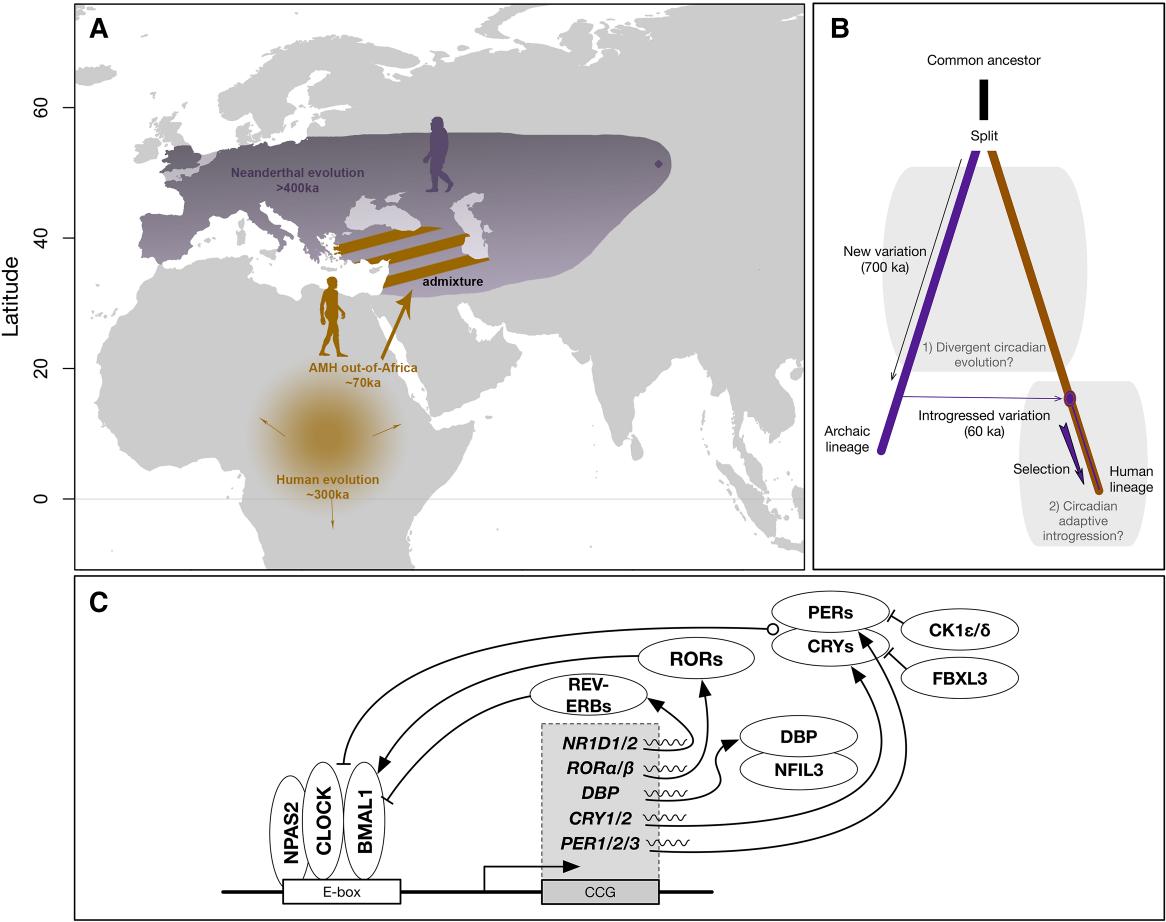
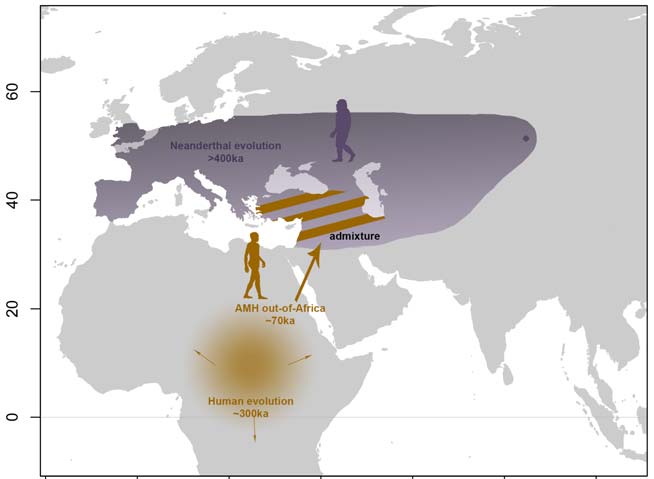
Archaic Introgression Shaped Human Circadian Traits
Velazquez-Arcelay K, Colbran LL, McArthur E, Brand CM, Rinker DC, Siemann JK, McMahon DG, Capra JA.*
Genome Biol Evol,
2023
Cell-Specific Transposable Element Gene Expression Analysis Identifies Associations with Systemic Lupus Erythematosus Phenotypes
Cutts Z, Patterson S, Maliskova L, Taylor KE, Ye C, Dall’Era M, Yazdany J, Criswell L, Fragiadakis GK, Langelier C, Capra JA, Sirota M^, Lanata CM.^*
bioRxiv,
2023
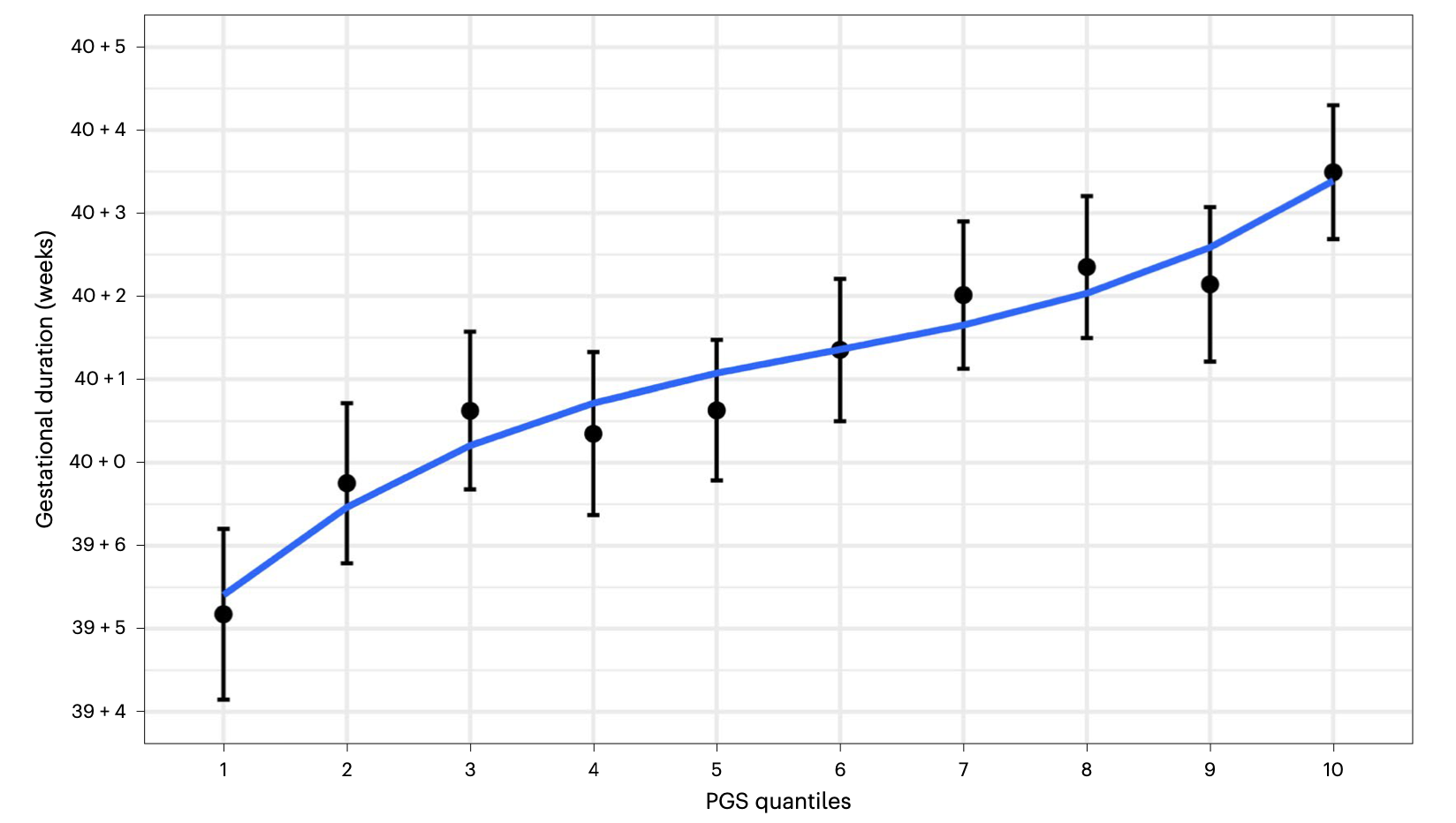
Associations with spontaneous and indicated preterm birth in a densely phenotyped EHR cohort
Costello JM^, Takasuka H^, Roger J, Yin O, Tang A, Oskotsky T, Sirota M*, Capra JA.*
medRxiv,
2023
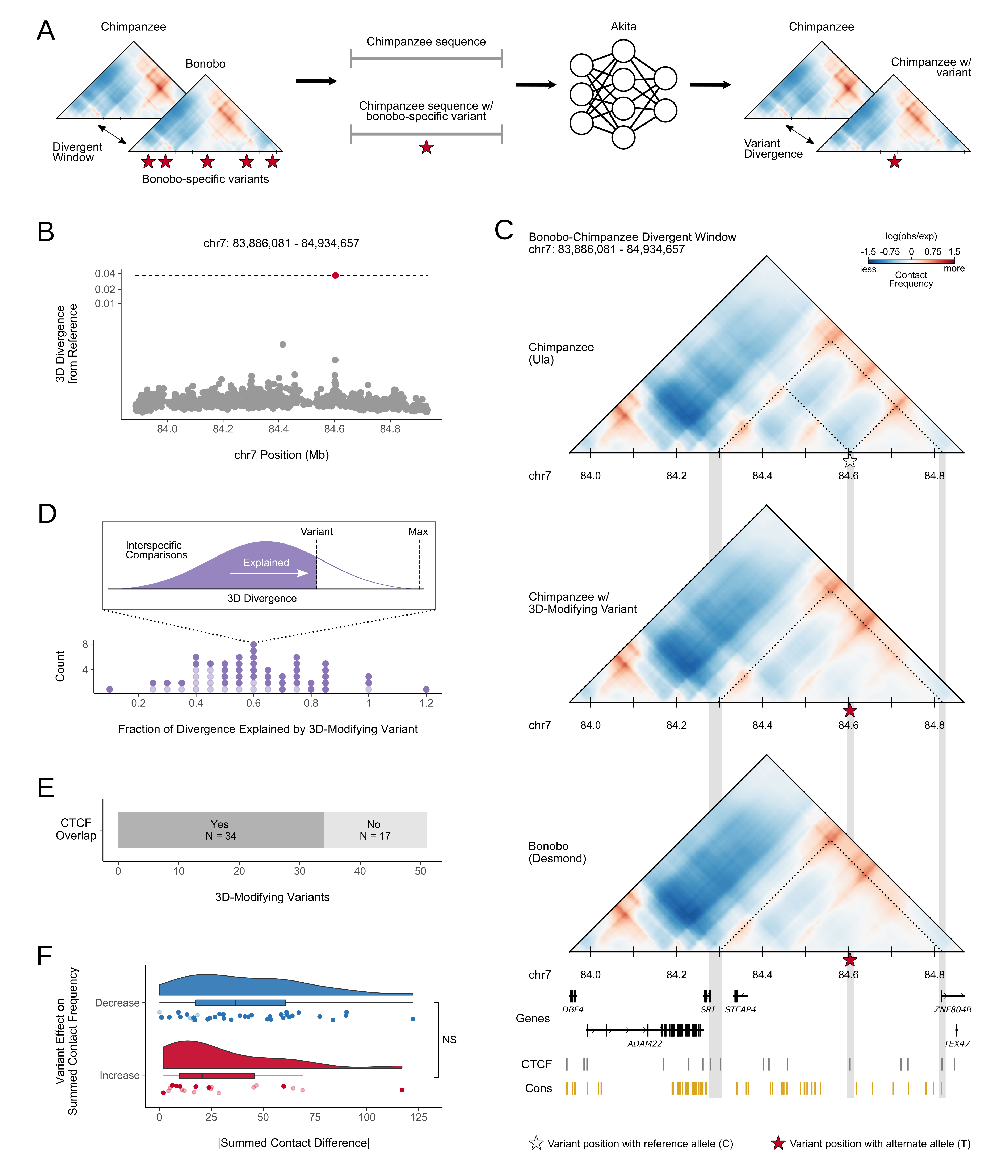
Sequence-based machine learning reveals 3D genome differences between bonobos and chimpanzees
Brand CM*, Kuang S, Gilbertson EN, McArthur E, Pollard KS, Webster TH, Capra JA.*
bioRxiv,
2023
Illuminating the Function of the Orphan Transporter, SLC22A10 in Humans and Other Primates
Yee SW, Ferrández-Peral L, Alentorn P, Fontsere C, Ceylan M, Koleske ML, Handin N, Artegoitia VM, Lara G, Chien HC, Zhou X, Dainat J, Zalevsky A, Sali A, Brand CM, Capra JA, Artursson P, Newman JW, Marques-Bonet T, Giacomini KM.*
Res Sq,
2023
Illuminating the Function of the Orphan Transporter, SLC22A10 in Humans and Other Primates
Yee SW, Ferrández-Peral L, Alentorn P, Fontsere C, Ceylan M, Koleske ML, Handin N, Artegoitia VM, Lara G, Chien HC, Zhou X, Dainat J, Zalevsky A, Sali A, Brand CM, Capra JA, Artursson P, Newman JW, Marques-Bonet T, Giacomini KM.*
bioRxiv,
2023
Integrating Computational Approaches to Predict the Effect of Genetic Variants on Protein Stability in Retinal Degenerative Disease
Grunin M^*, Palmer E^, de Jong S^*, Jin B, Rinker D, Moth C, Capra JA, Haines JL^, Bush WS^, den Hollander AI.^
Adv Exp Med Biol,
2023
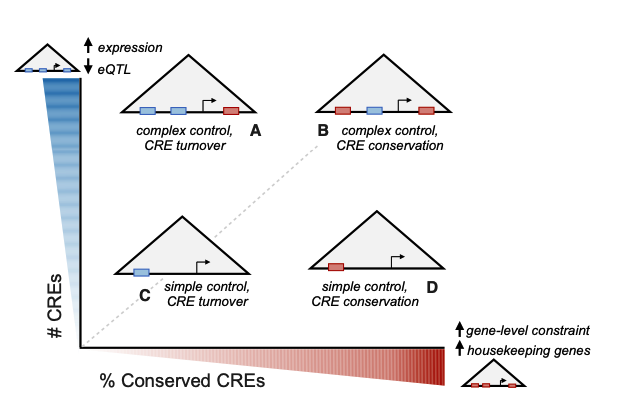
Cis-regulatory Landscape Size, Constraint, and Tissue Specificity Associate with Gene Function and Expression
Benton ML*, Ruderfer DM, Capra JA.*
Genome Biol Evol,
2023
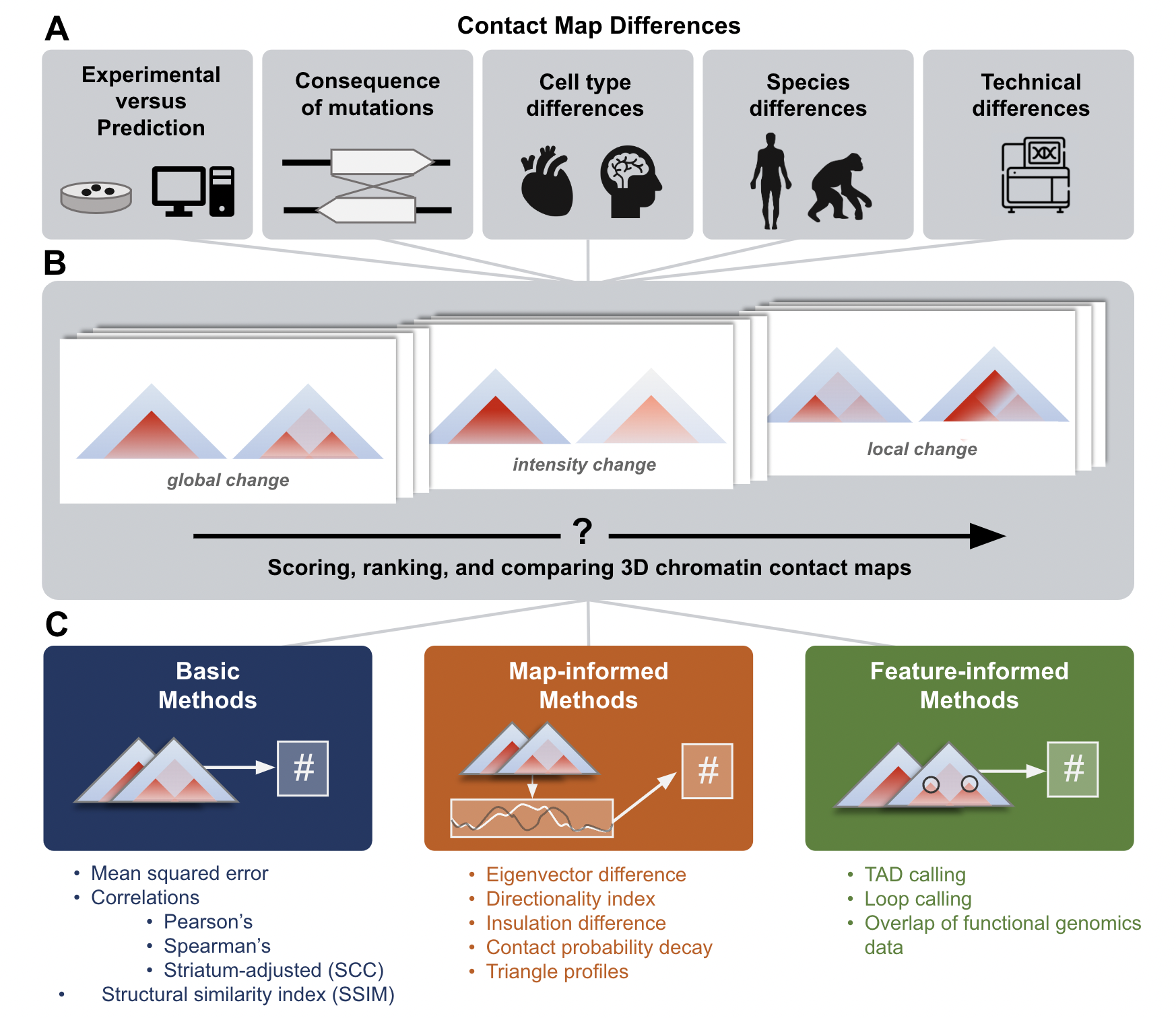
Comparing chromatin contact maps at scale: methods and insights
Gunsalus LM^, McArthur E^, Gjoni K, Kuang S, Pittman M, Capra JA*, Pollard KS.*
Res Sq,
2023
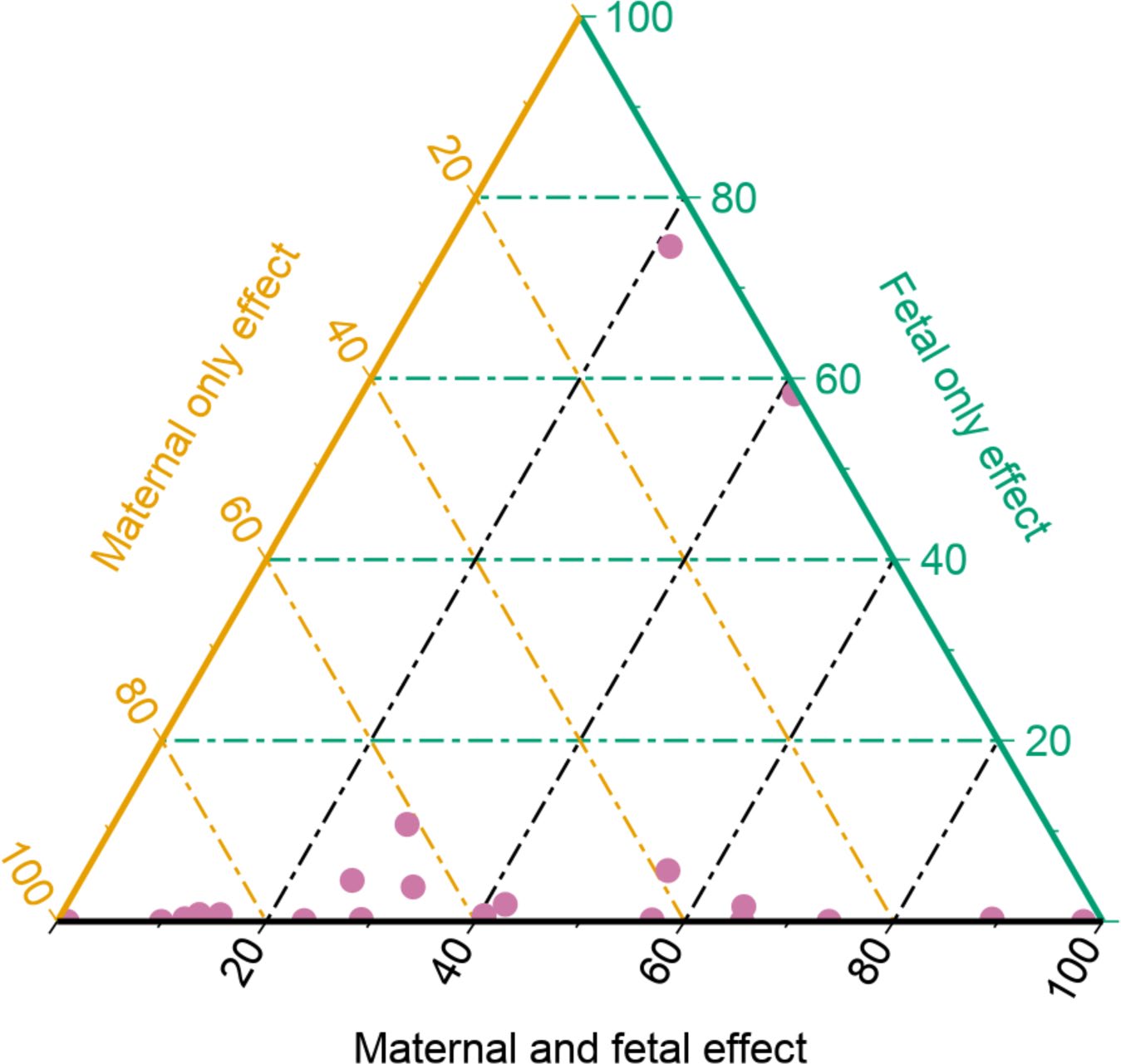
Author Correction: Genetic effects on the timing of parturition and links to fetal birth weight
Solé-Navais P*, Flatley C, Steinthorsdottir V, Vaudel M, Juodakis J, Chen J, Laisk T, LaBella AL, Westergaard D, Bacelis J, Brumpton B, Skotte L, Borges MC, Helgeland Ø, Mahajan A, Wielscher M, Lin F, Briggs C, Wang CA, Moen GH, Beaumont RN, Bradfield JP, Abraham A, Thorleifsson G, Gabrielsen ME, Ostrowski SR, Modzelewska D, Nohr EA, Hypponen E, Srivastava A, Talbot O, Allard C, Williams SM, Menon R, Shields BM, Sveinbjornsson G, Xu H, Melbye M, Lowe W Jr, Bouchard L, Oken E, Pedersen OB, Gudbjartsson DF, Erikstrup C, Sørensen E; Early Growth Genetics Consortium; Estonian Biobank Research Team; Danish Blood Donor Study Genomic Consortium; Lie RT, Teramo K, Hallman M, Juliusdottir T, Hakonarson H, Ullum H, Hattersley AT, Sletner L, Merialdi M, Rifas-Shiman SL, Steingrimsdottir T, Scholtens D, Power C, West J, Nyegaard M, Capra JA, Skogholt AH, Magnus P, Andreassen OA, Thorsteinsdottir U, Grant SFA, Qvigstad E, Pennell CE, Hivert MF, Hayes GM, Jarvelin MR, McCarthy MI, Lawlor DA, Nielsen HS, Mägi R, Rokas A, Hveem K, Stefansson K, Feenstra B, Njolstad P, Muglia LJ, Freathy RM, Johansson S, Zhang G, Jacobsson B.*
Nat Genet,
2023
High-throughput functional mapping of variants in an arrhythmia gene, KCNE1, reveals novel biology
Muhammad A, Calandranis ME, Li B, Yang T, Blackwell DJ, Harvey ML, Smith JE, Chew AE, Capra JA, Matreyek KA, Fowler DM, Roden DM^, Glazer AM.^*
bioRxiv,
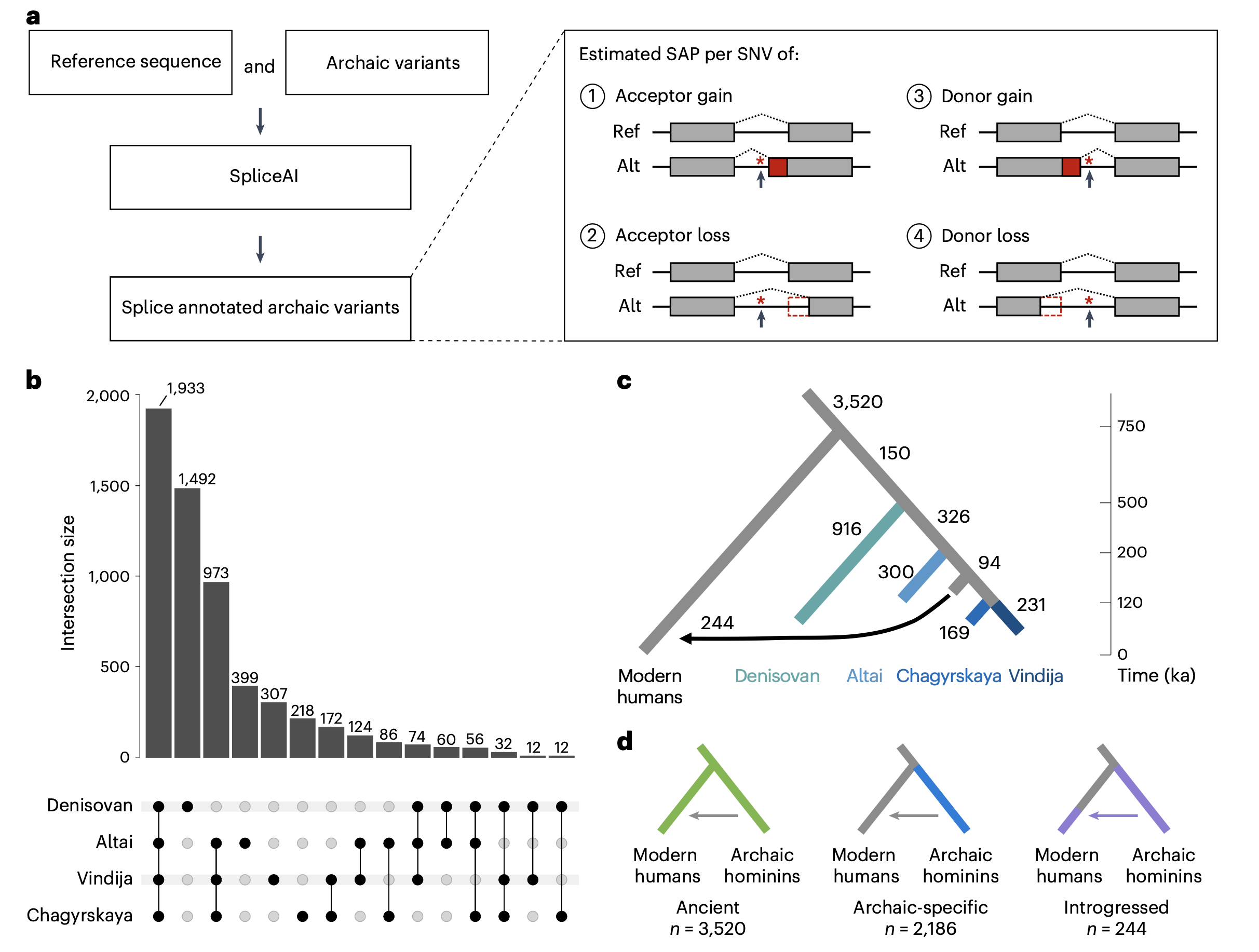
Resurrecting the alternative splicing landscape of archaic hominins using machine learning
Brand CM, Colbran LL, Capra JA*
Nature Ecology & Evolution,
Comparing chromatin contact maps at scale: methods and insights
Gunsalus LM^, McArthur E^, Gjoni K, Kuang S, Pittman M, Capra JA*, Pollard KS.*
bioRxiv,
2023
Genetic effects on the timing of parturition and links to fetal birth weight
Solé-Navais P*, Flatley C, Steinthorsdottir V, Vaudel M, Juodakis J, Chen J, Laisk T, LaBella AL, Westergaard D, Bacelis J, Brumpton B, Skotte L, Borges MC, Helgeland Ø, Mahajan A, Wielscher M, Lin F, Briggs C, Wang CA, Moen GH, Beaumont RN, Bradfield JP, Abraham A, Thorleifsson G, Gabrielsen ME, Ostrowski SR, Modzelewska D, Nohr EA, Hypponen E, Srivastava A, Talbot O, Allard C, Williams SM, Menon R, Shields BM, Sveinbjornsson G, Xu H, Melbye M, Lowe W Jr, Bouchard L, Oken E, Pedersen OB, Gudbjartsson DF, Erikstrup C, Sørensen E; Early Growth Genetics Consortium; Estonian Biobank Research Team; Danish Blood Donor Study Genomic Consortium; Lie RT, Teramo K, Hallman M, Juliusdottir T, Hakonarson H, Ullum H, Hattersley AT, Sletner L, Merialdi M, Rifas-Shiman SL, Steingrimsdottir T, Scholtens D, Power C, West J, Nyegaard M, Capra JA, Skogholt AH, Magnus P, Andreassen OA, Thorsteinsdottir U, Grant SFA, Qvigstad E, Pennell CE, Hivert MF, Hayes GM, Jarvelin MR, McCarthy MI, Lawlor DA, Nielsen HS, Mägi R, Rokas A, Hveem K, Stefansson K, Feenstra B, Njolstad P, Muglia LJ, Freathy RM, Johansson S, Zhang G, Jacobsson B.*
Nat Genet,
2023
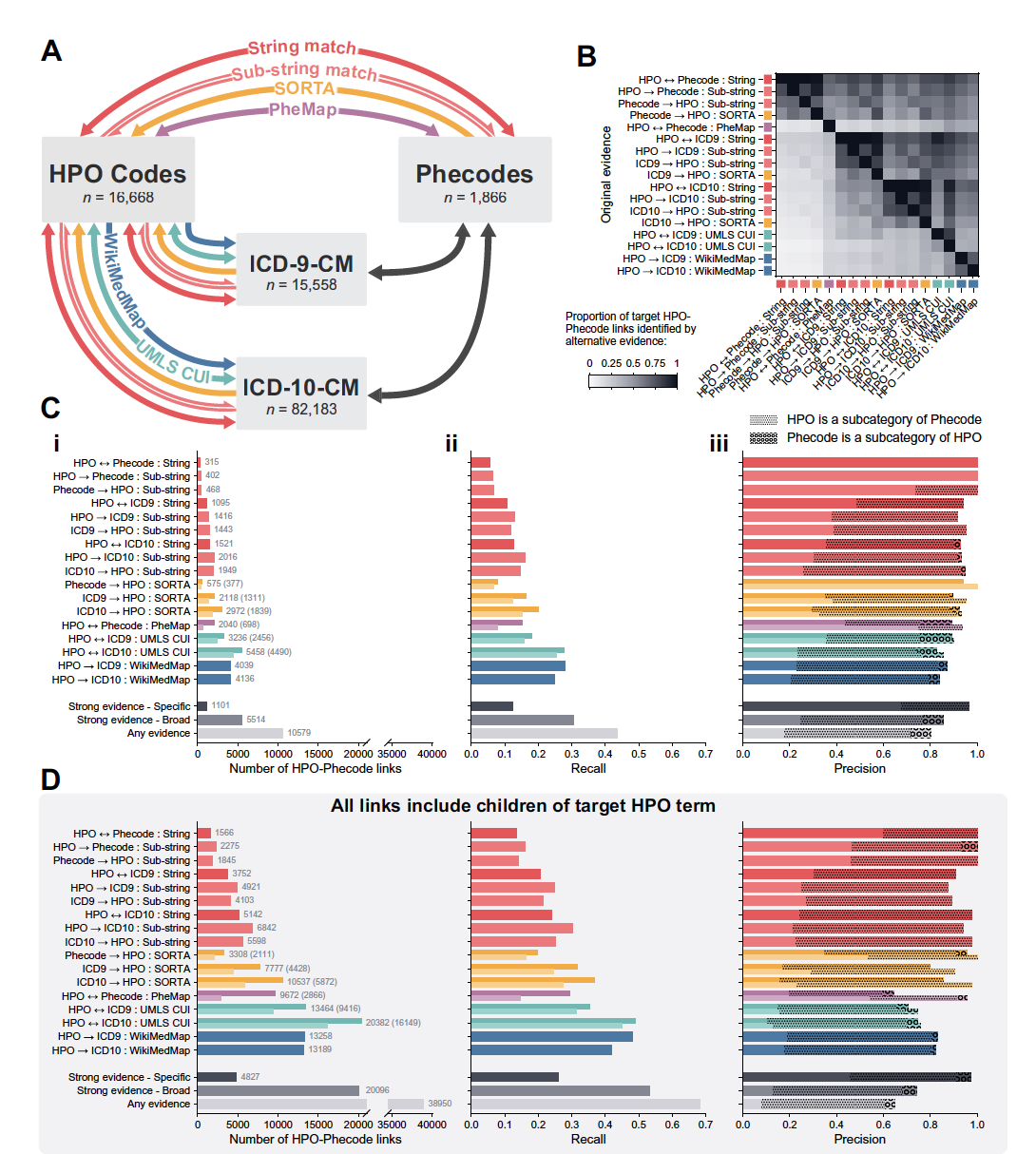
Linking rare and common disease vocabularies by mapping between the human phenotype ontology and phecodes
McArthur E*, Bastarache L, Capra JA.
JAMIA Open,
2023
Human gene regulatory evolution is driven by the divergence of regulatory element function in both cis and trans
Hansen T^, Fong S^, Capra JA*, Hodges E.*
bioRxiv,
2023
Archaic Introgression Shaped Human Circadian Traits
Velazquez-Arcelay K, Colbran LL, McArthur E, Brand C, Rinker D, Siemann J, McMahon D, Capra JA.*
bioRxiv,
2023
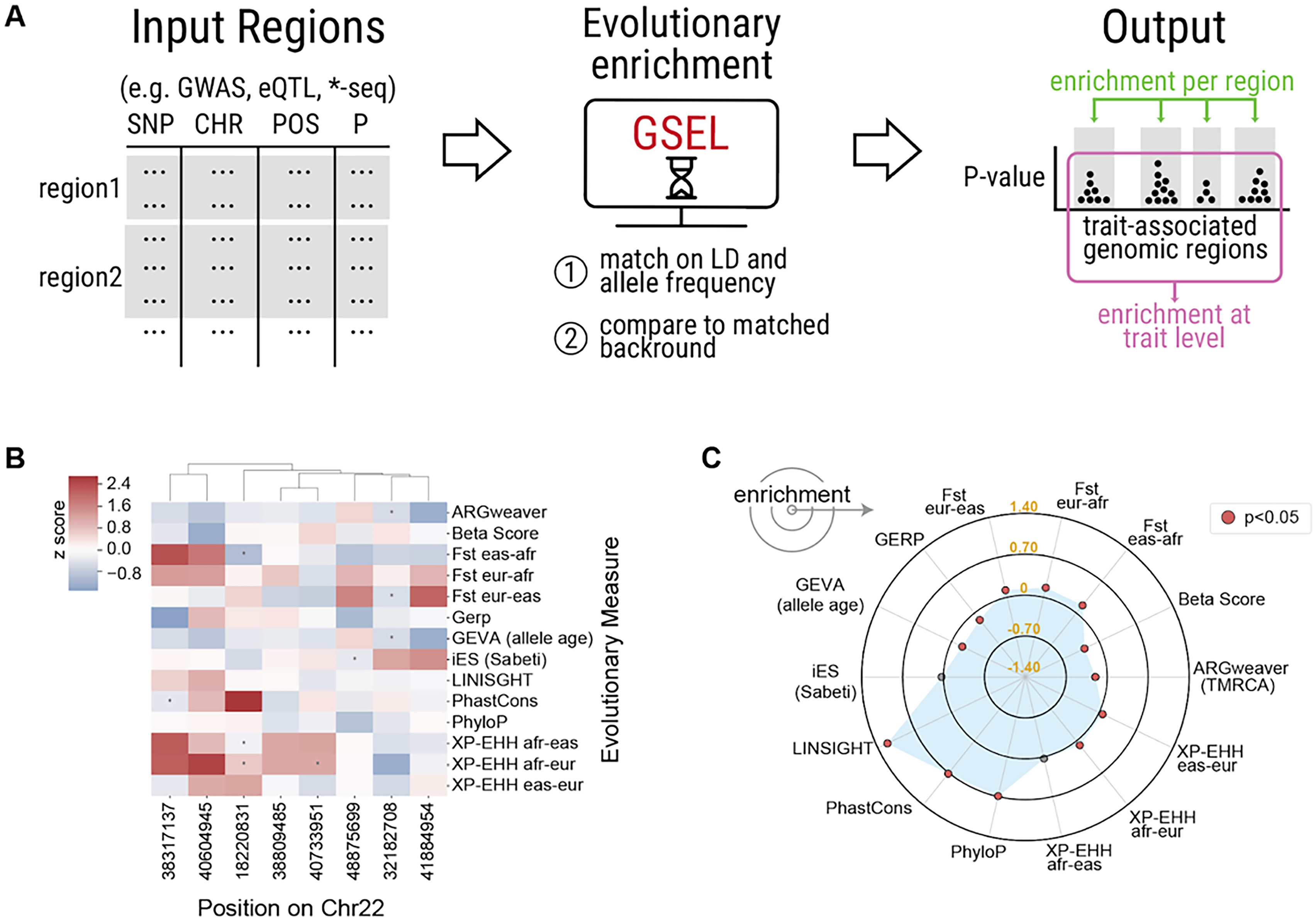
GSEL: a fast, flexible python package for detecting signatures of diverse evolutionary forces on genomic regions
Abraham A*, Labella AL, Benton ML, Rokas A, Capra JA.*
Bioinformatics,
2023
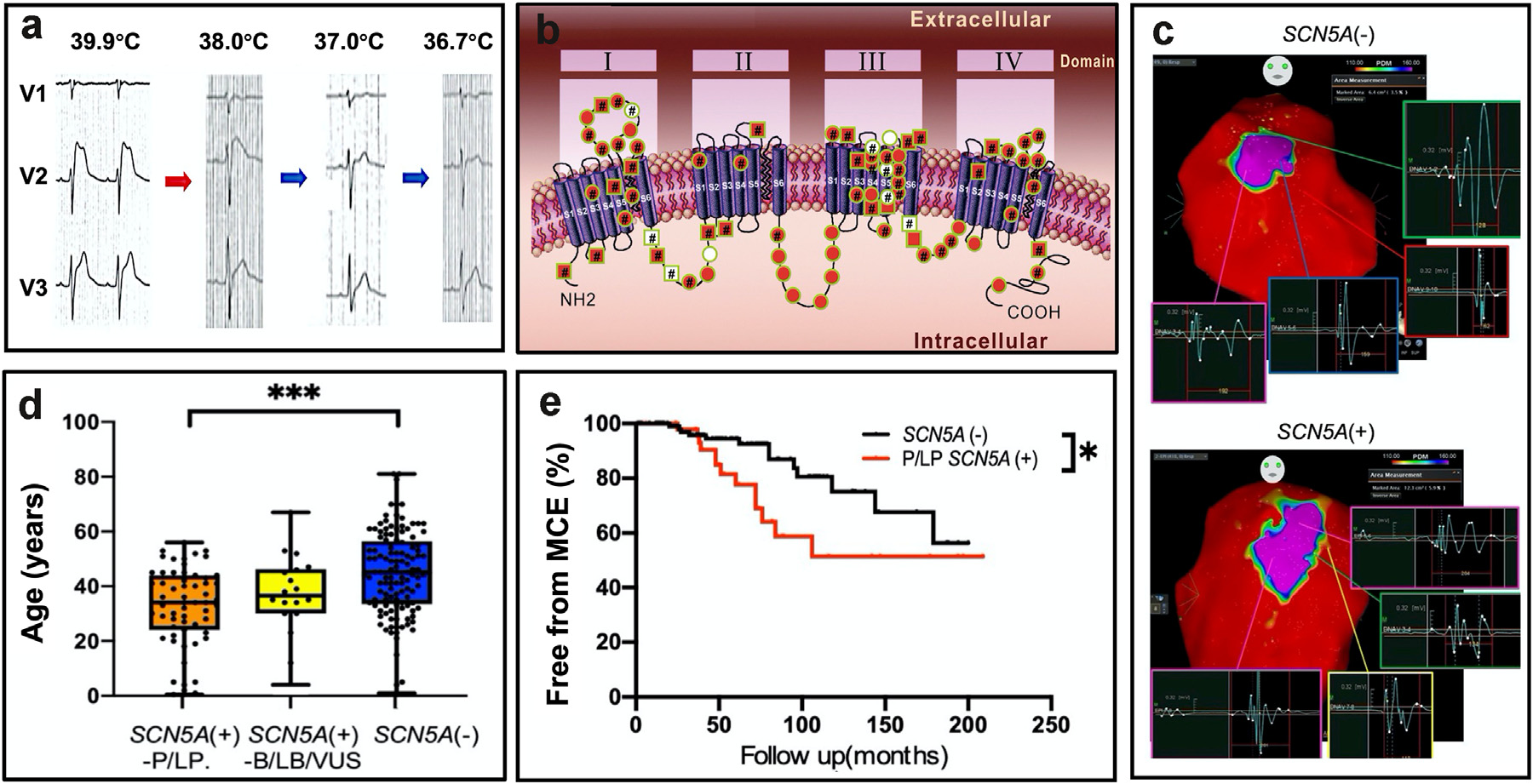
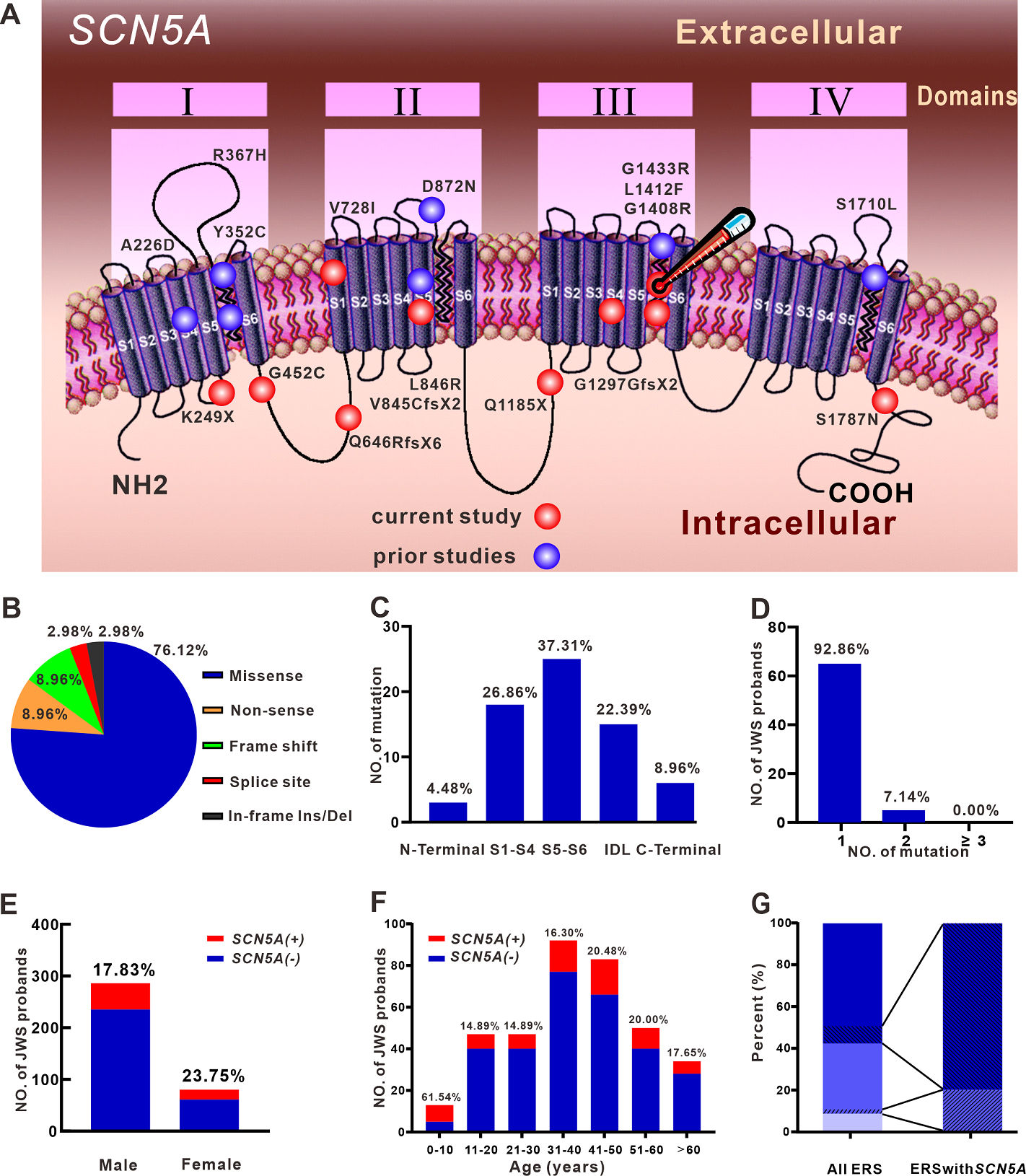
Clinical characteristics and electrophysiologic properties of SCN5A variants in fever-induced Brugada syndrome
Chen GX^, Barajas-Martínez H^, Ciconte G^, Wu CI^, Monasky MM, Xia H, Li B, Capra JA, Guo K, Zhang ZH, Chen X, Yang B, Jiang H, Tse G, Mak CM, Aizawa Y, Gollob MH, Antzelevitch C, Wilde AAM5, Pappone C#, Hu D.#*
EBioMedicine,
2022
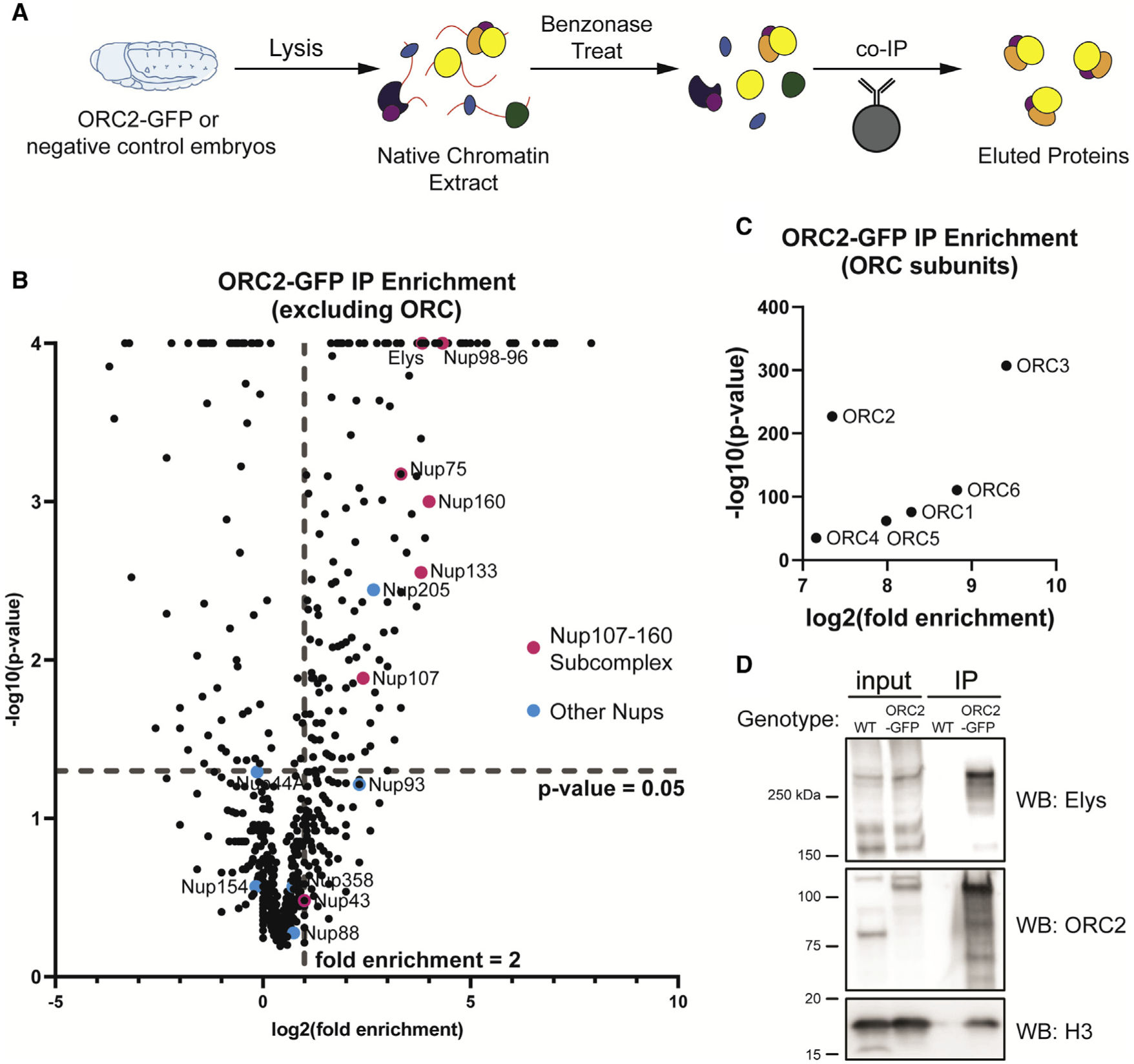
Nucleoporins facilitate ORC loading onto chromatin
Richards L, Lord CL, Benton ML, Capra JA, Nordman JT.*
Cell Rep,
2022
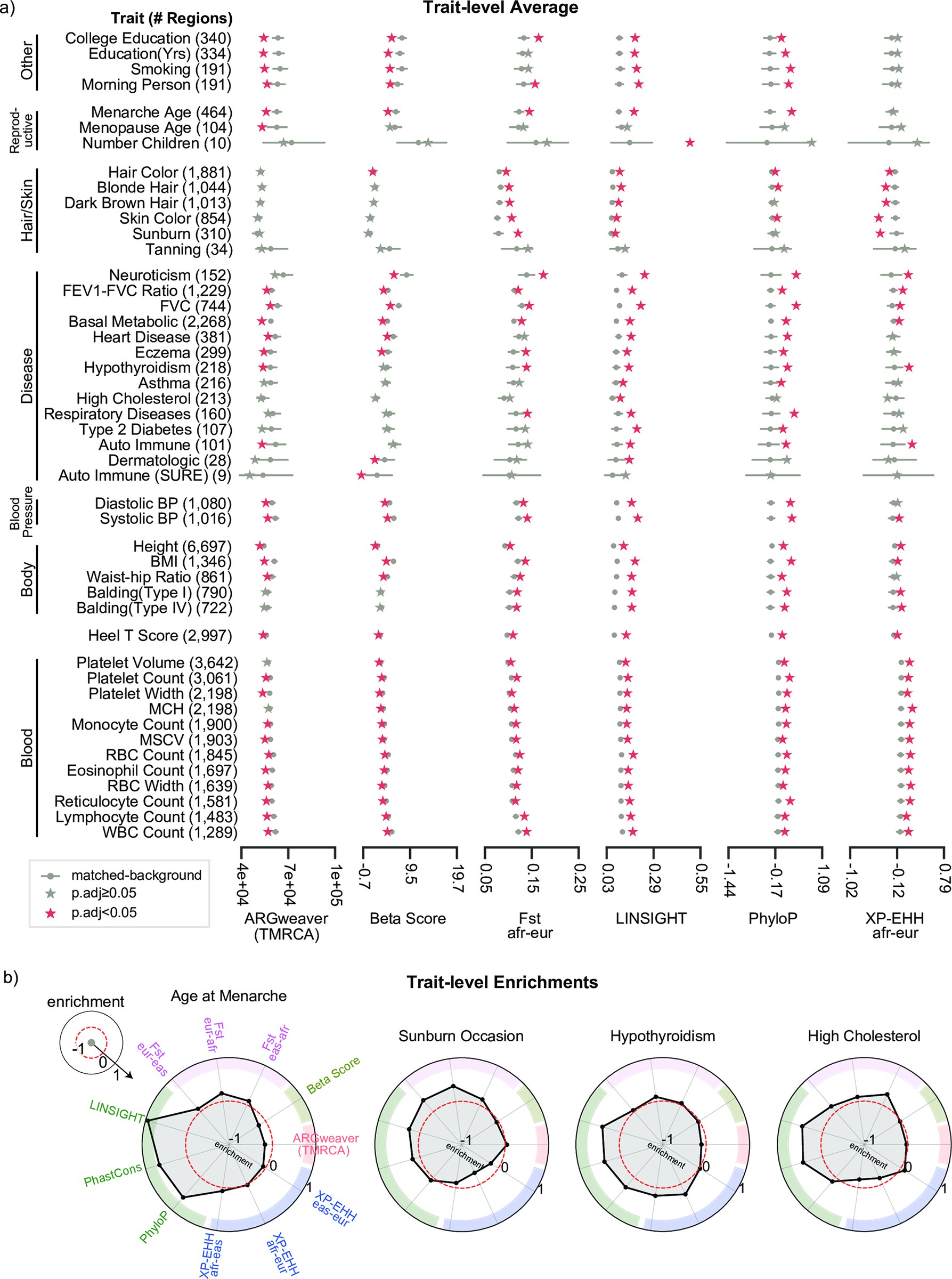
Mosaic patterns of selection in genomic regions associated with diverse human traits
Abraham A^, LaBella AL^, Capra JA*, Rokas A.*
PLoS Genet,
2022
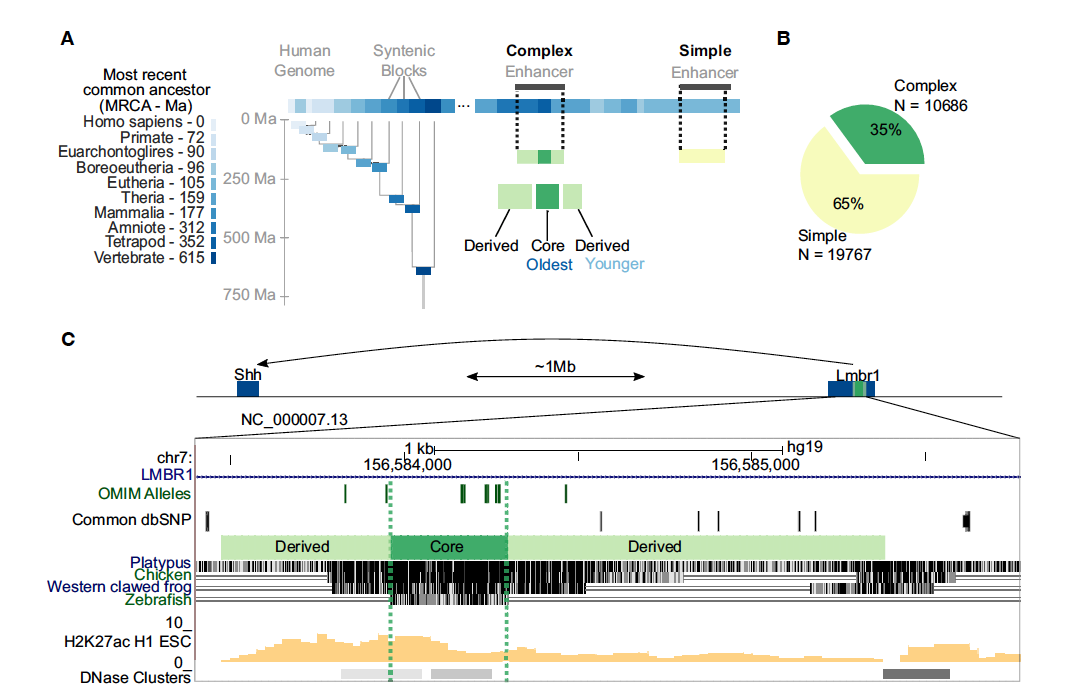
Function and Constraint in Enhancer Sequences with Multiple Evolutionary Origins
Fong SL, Capra JA.*
Genome Biol Evol,
2022
Dense phenotyping from electronic health records enables machine learning-based prediction of preterm birth
Abraham A, Le B, Kosti I, Straub P, Velez-Edwards DR, Davis LK, Newton JM, Muglia LJ, Rokas A, Bejan CA, Sirota M, Capra JA.*
BMC Med,
2022
The immune deficiency and c-Jun N-terminal kinase pathways drive the functional integration of the immune and circulatory systems of mosquitoes
Yan Y^, Sigle LT^, Rinker DC, Estévez-Lao TY, Capra JA, Hillyer JF.*
Open Biol,
2022
Personalized structural biology reveals the molecular mechanisms underlying heterogeneous epileptic phenotypes caused by de novo KCNC2 variants
Mukherjee S, Cassini TA^, Hu N, Yang T^, Li B^, Shen W, Moth CW, Rinker DC, Sheehan JH, Cogan JD; Undiagnosed Diseases Network; Newman JH, Hamid R, Macdonald RL, Roden DM, Meiler J, Kuenze G*, Phillips JA*, Capra JA.*
HGG Adv,
2022
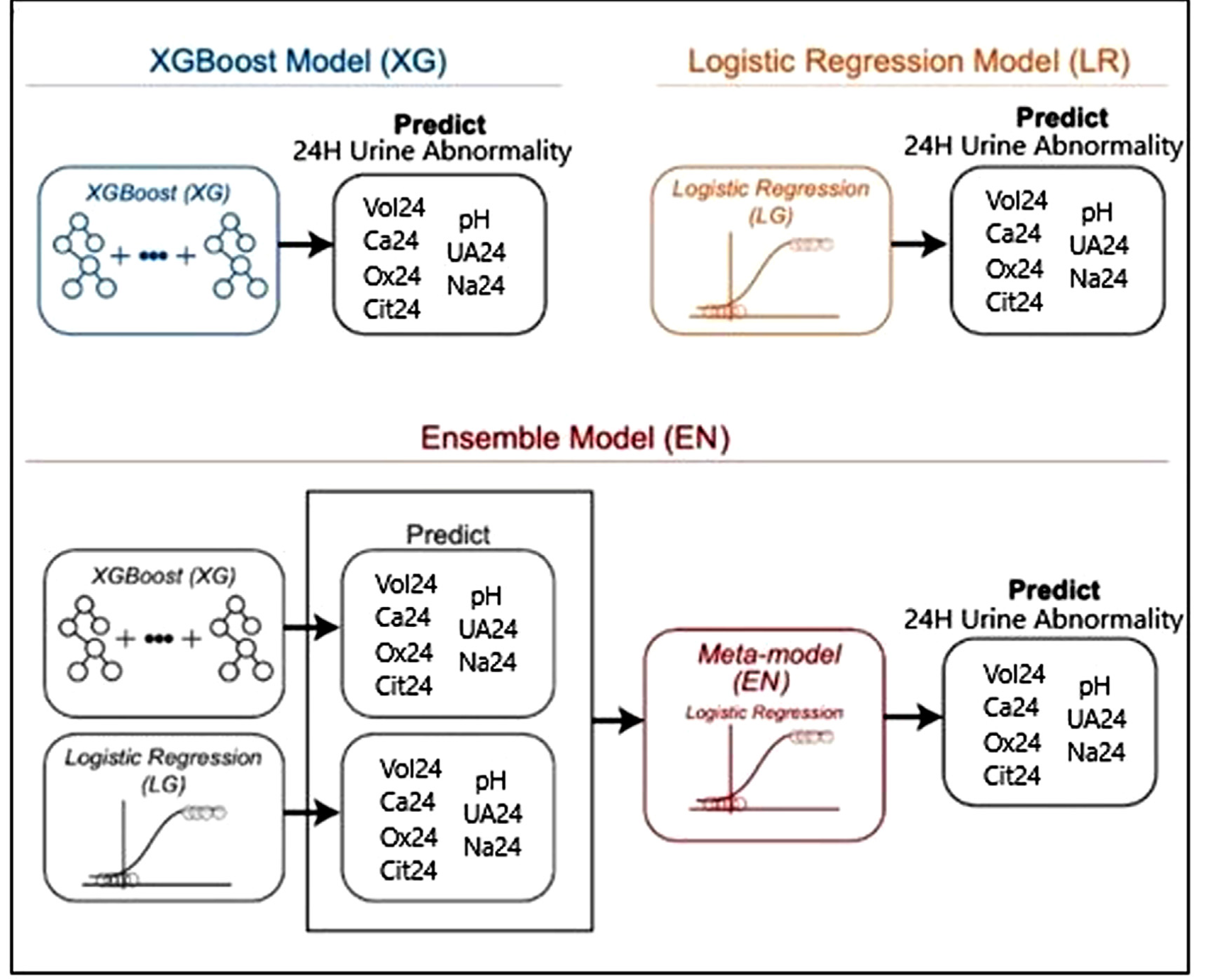
Machine Learning Models to Predict 24 Hour Urinary Abnormalities for Kidney Stone Disease
Kavoussi NL*, Floyd C, Abraham A, Sui W, Bejan C, Capra JA, Hsi R.
Urology,
2022
Microbiome-associated human genetic variants impact phenome-wide disease risk
Markowitz RHG, LaBella AL, Shi M, Rokas A, Capra JA, Ferguson JF, Mosley JD, Bordenstein SR.*
Proc Natl Acad Sci U S A,
2022
Genome-wide association study of musical beat synchronization demonstrates high polygenicity
Niarchou M*, Gustavson DE, Sathirapongsasuti JF, Anglada-Tort M, Eising E, Bell E, McArthur E, Straub P; 23andMe Research Team; McAuley JD, Capra JA, Ullén F, Creanza N, Mosing MA, Hinds DA, Davis LK#*, Jacoby N#, Gordon RL.#*
Nat Hum Behav,
2022
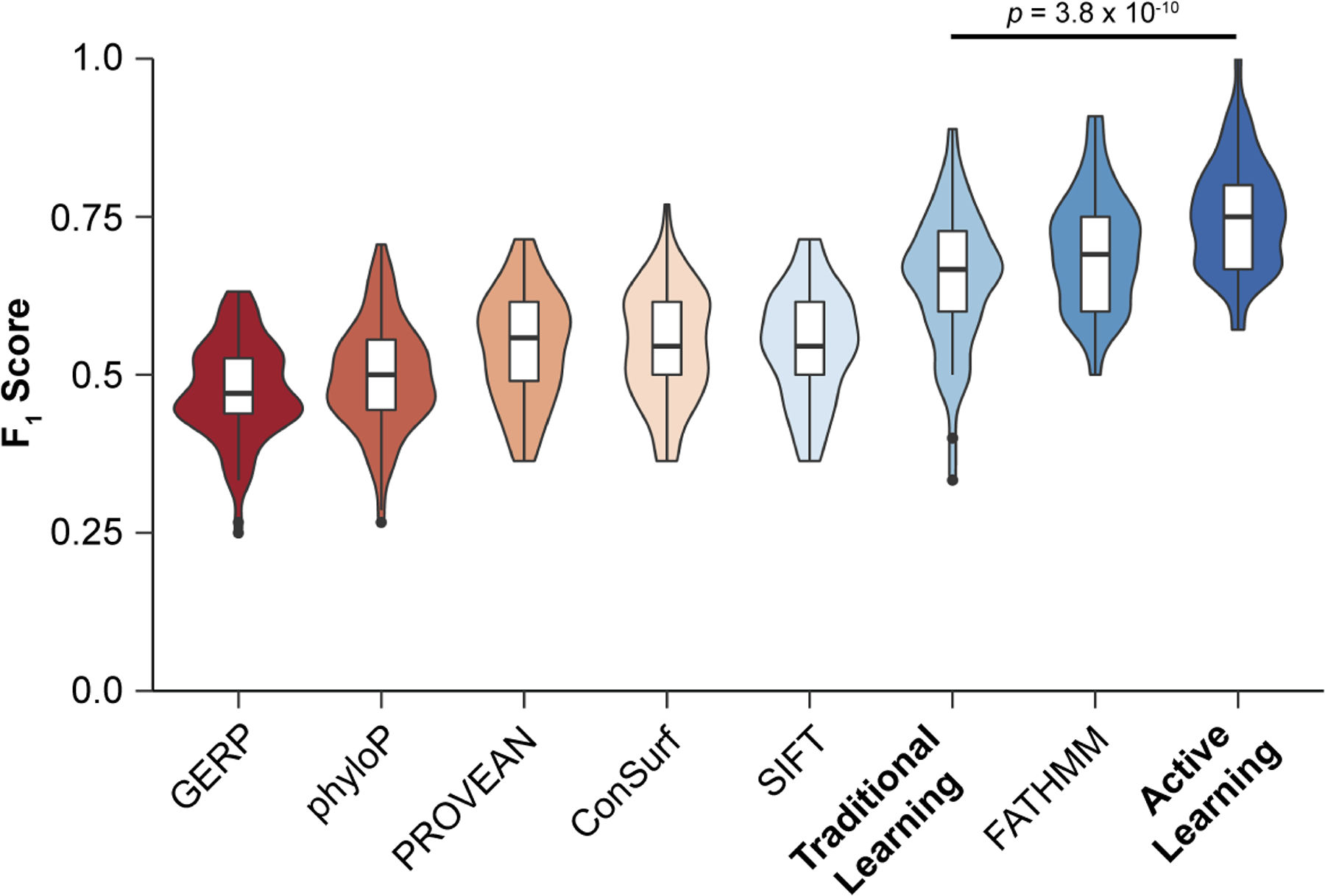
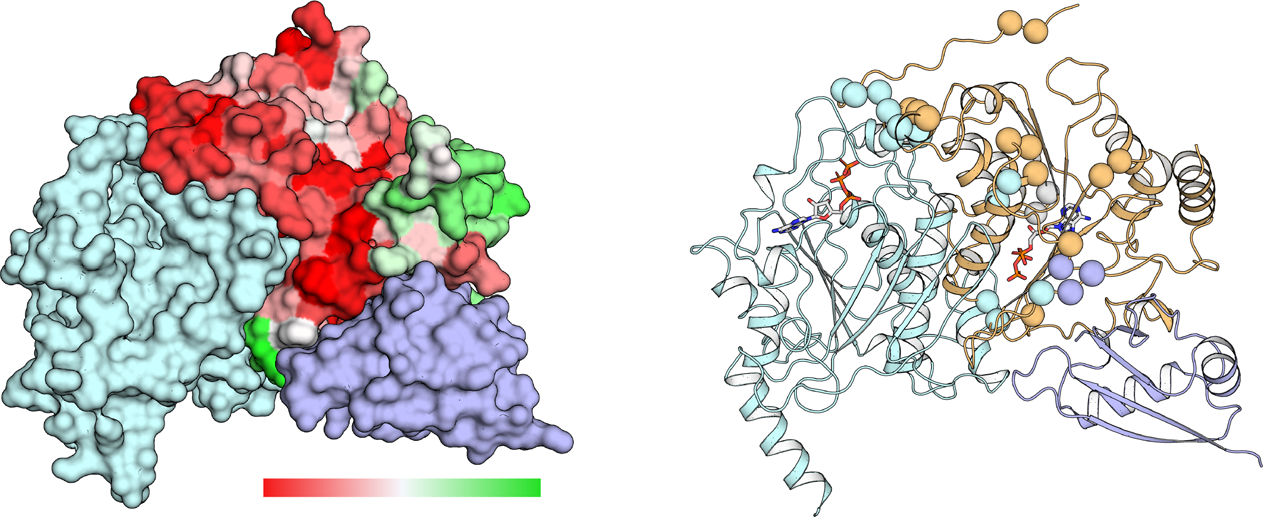
An Active Learning Framework Improves Tumor Variant Interpretation
Blee AM^, Li B^, Pecen T, Meiler J, Nagel ZD, Capra JA*, Chazin WJ.*
Cancer Res,
2022
The 3D mutational constraint on amino acid sites in the human proteome
Li B*, Roden DM, Capra JA.*
Nat Commun,
2022
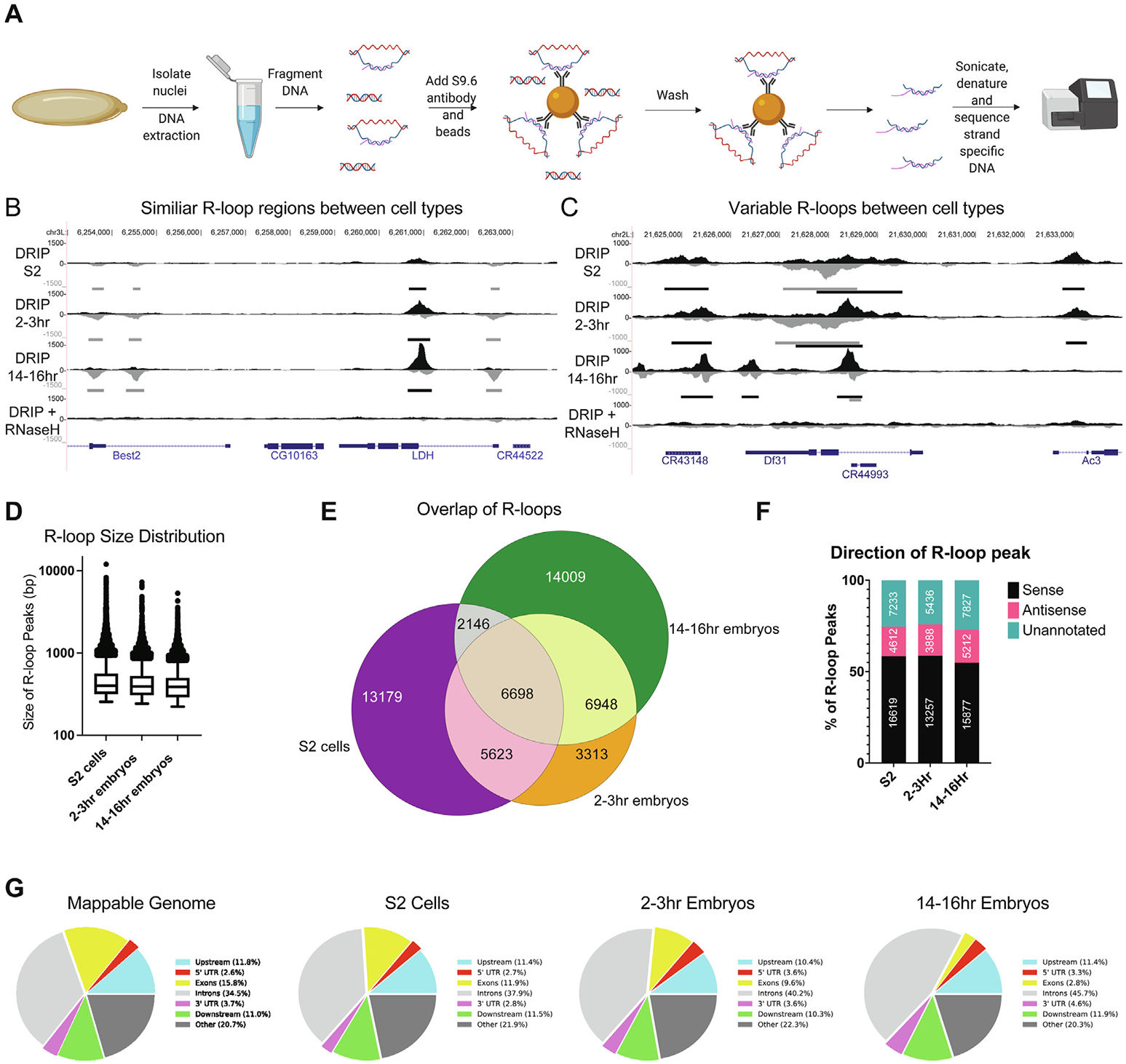
R-loop Mapping and Characterization During Drosophila Embryogenesis Reveals Developmental Plasticity in R-loop Signatures
Munden A, Benton ML, Capra JA, Nordman JT.*
J Mol Biol,
2022
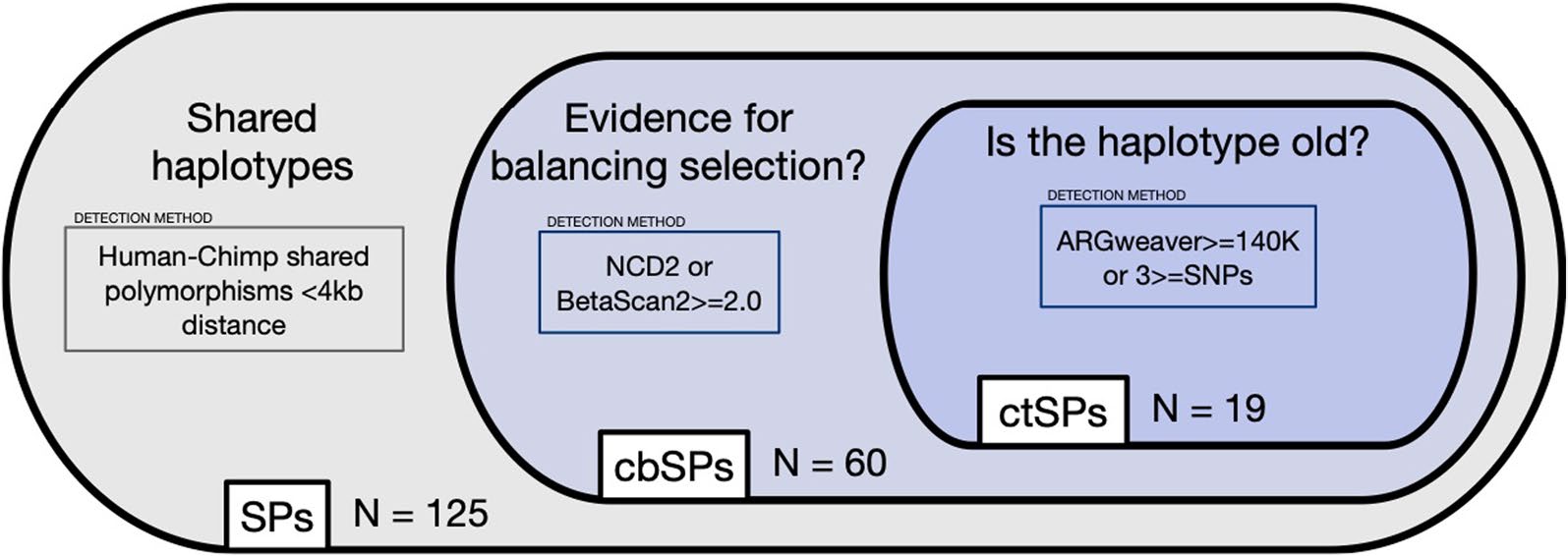
Diverse functions associate with non-coding polymorphisms shared between humans and chimpanzees
Velazquez-Arcelay K, Benton ML, Capra JA.*
BMC Ecol Evol,
2022
Vascular alterations impede fragile tolerance to pregnancy in type 1 diabetes
McNew KL, Abraham A, Sack DE, Smart CD, Pettway YD, Falk AC, Lister RL, Faucon AB, Bejan CA, Capra JA, Aronoff DM, Boyd KL, Moore DJ.*
F S Sci,
2022
Integration of Protein Structure and Population-Scale DNA Sequence Data for Disease Gene Discovery and Variant Interpretation
Li B, Jin B, Capra JA, Bush WS.*
Annu Rev Biomed Data Sci,
2022
Predicting archaic hominin phenotypes from genomic data
Brand CM, Colbran LL, Capra JA*
Annual Review of Genetics and Human Genomics,
2022
An association test of the spatial distribution of rare missense variants within protein structures identifies Alzheimer’s disease-related patterns
Jin B, Capra JA, Benchek P, Wheeler N, Naj AC, Hamilton-Nelson KL, Farrell JJ, Leung YY, Kunkle B, Vadarajan B, Schellenberg GD, Mayeux R, Wang LS, Farrer LA, Pericak-Vance MA, Martin ER, Haines JL, Crawford DC, Bush WS.*
Genome Res,
2022
Tracing the Evolution of Human Gene Regulation and Its Association with Shifts in Environment
Colbran LL*, Johnson MR, Mathieson I, Capra JA.*
Genome Biol Evol,
2021
Distinct Features of Probands With Early Repolarization and Brugada Syndromes Carrying SCN5A Pathogenic Variants
Zhang ZH^, Barajas-Martínez H^, Xia H, Li B, Capra JA, Clatot J, Chen GX, Chen X, Yang B, Jiang H, Tse G, Aizawa Y, Gollob MH, Scheinman M, Antzelevitch C, Hu D.*
J Am Coll Cardiol,
2021
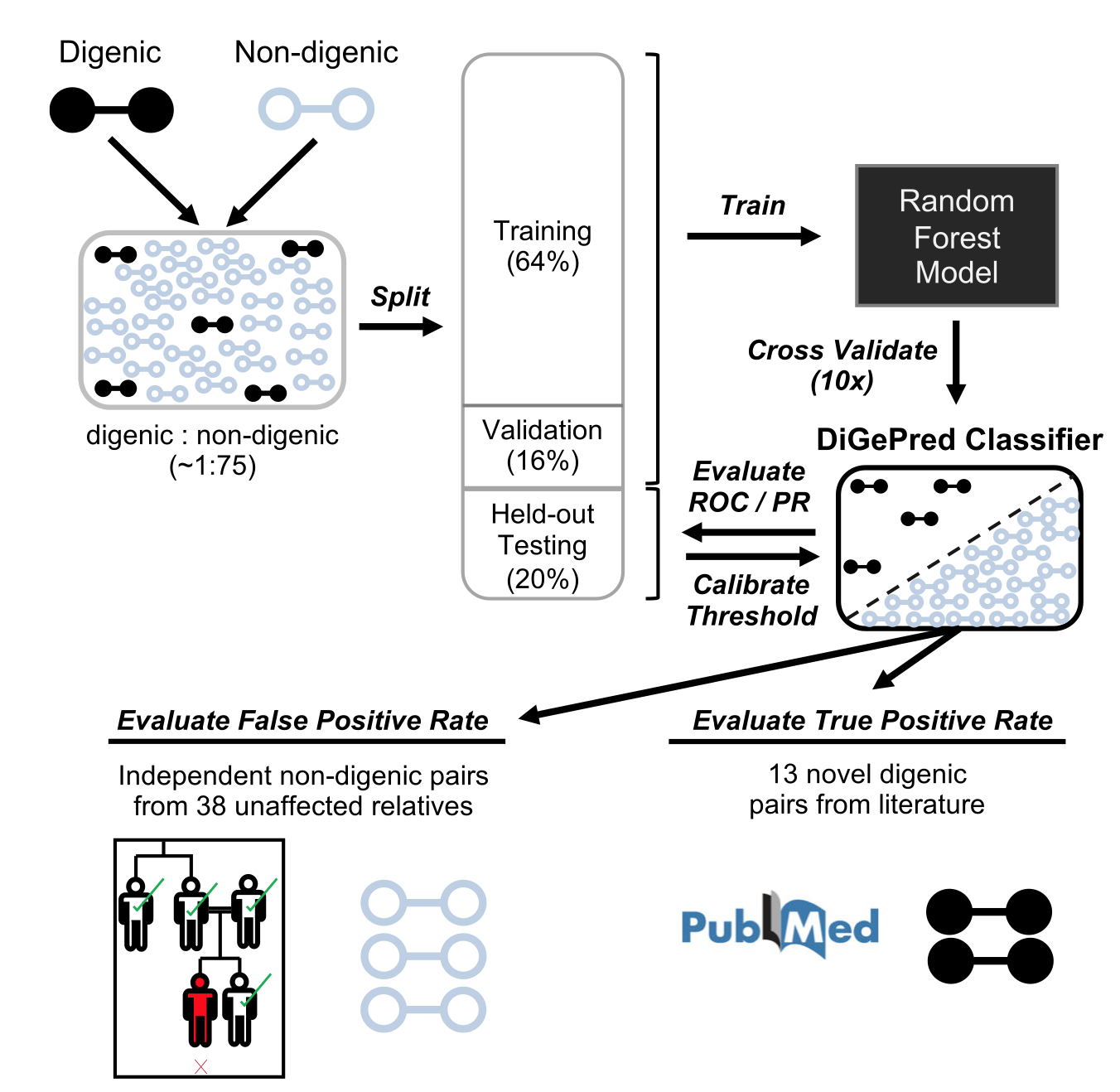
Identifying digenic disease genes via machine learning in the Undiagnosed Diseases Network
Mukherjee S, Cogan JD, Newman JH, Phillips JA III, Hamid R Undiagnosed Diseases Network, Meiler J*, Capra JA*
American Journal of Human Genetics (AJHG),
2021
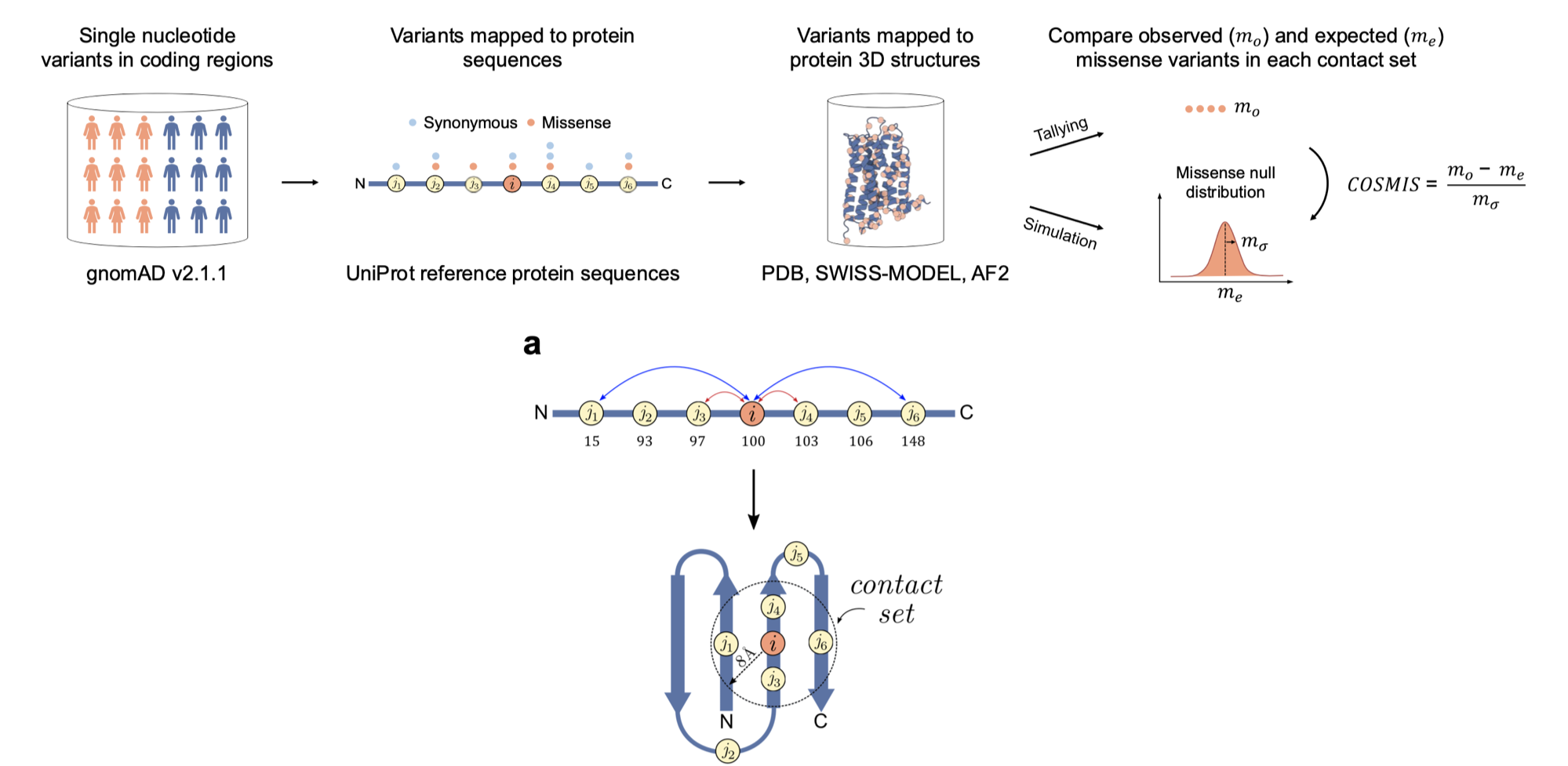
The 3D spatial constraint on 6.1 million amino acid sites in the human proteome
Li B, Roden DM, Capra JA*
bioRxiv,
Access the paper
- Biorxiv Preprint: 460390
- Full Text
- GitHub Repositories:
https://github.com/CapraLab/cosmis (COSMIS is a new framework for quantification of the constraint on protein-coding genetic variation in 3D spatial neighborhoods. It leverages recent advances in computational structure prediction, large-scale sequencing data from gnomAD, and a mutation-spectrum-aware statistical model.),
http://cosmis-app.herokuapp.com/
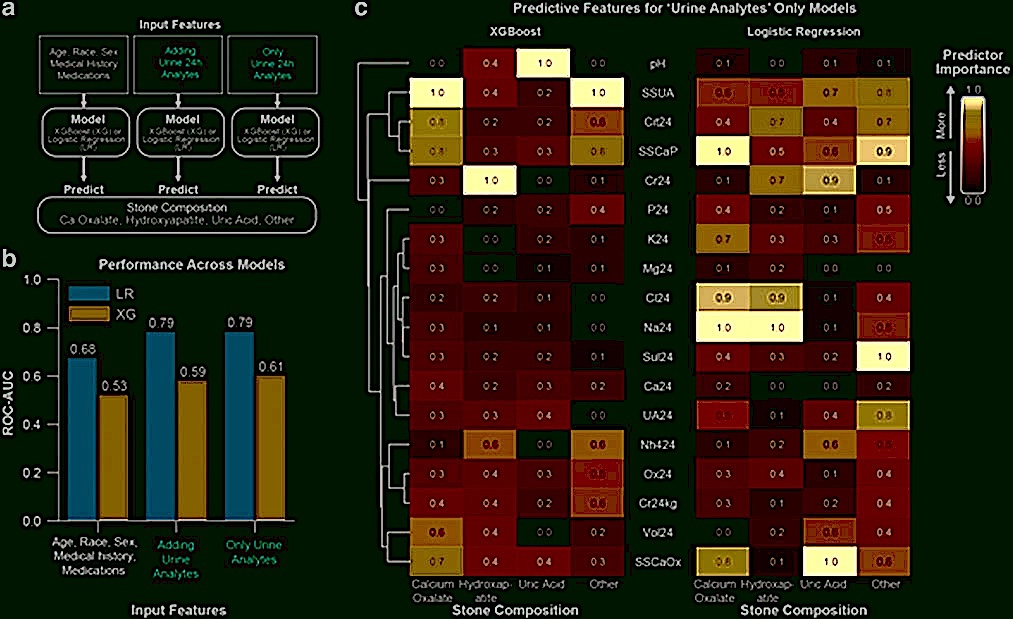
Machine Learning Prediction of Kidney Stone Composition Using Electronic Health Record-Derived Features
Abraham A^, Kavoussi NL^*, Sui W, Bejan C, Capra JA#, Hsi R.#
J Endourol,
2021
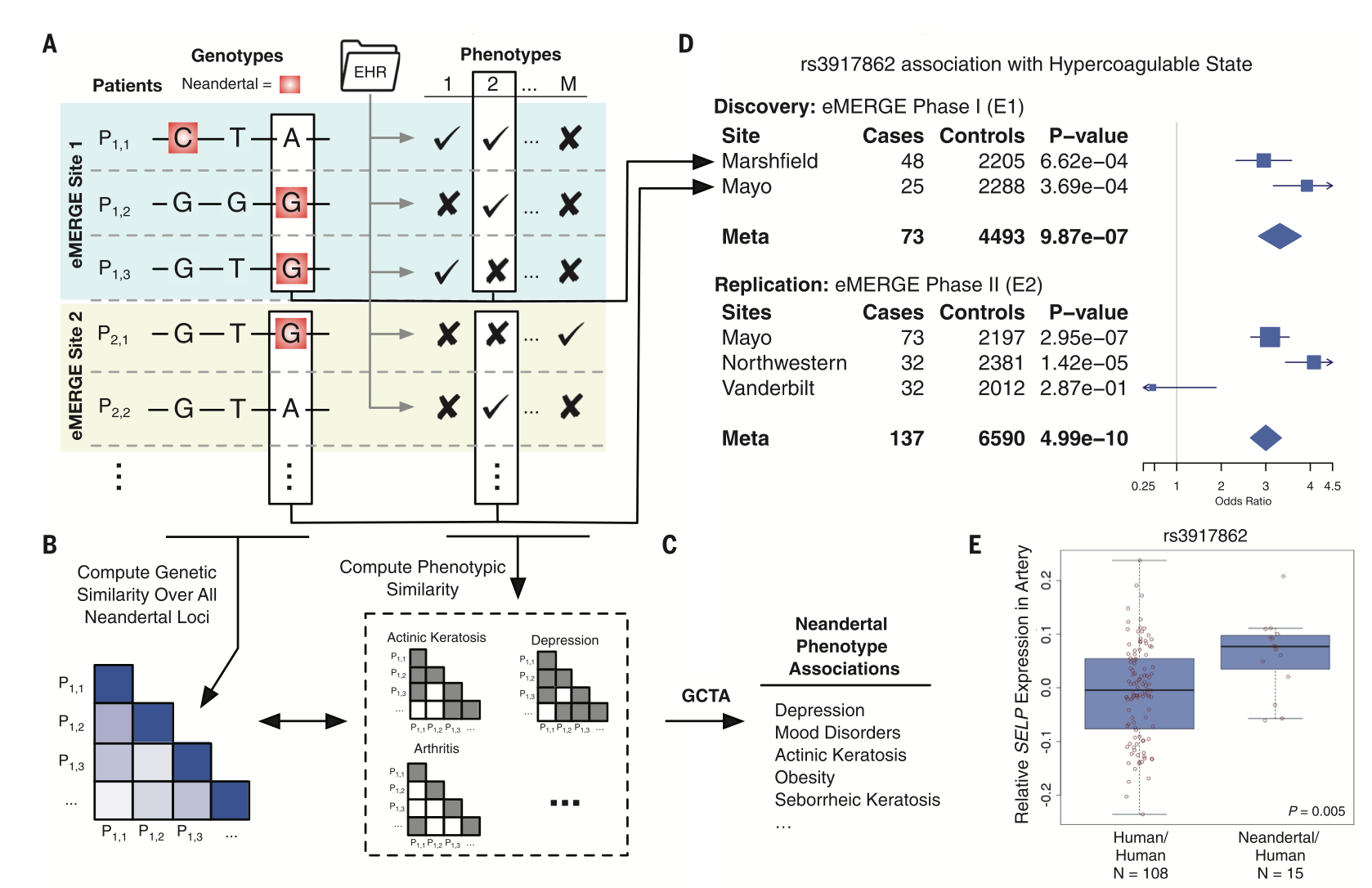
Quantifying the contribution of Neanderthal introgression to the heritability of complex traits
McArthur E, Rinker DC, Capra JA.*
Nat Commun,
2021
A Multitask Deep-Learning Method for Predicting Membrane Associations and Secondary Structures of Proteins
Li B, Mendenhall J, Capra JA, Meiler J.*
J Proteome Res,
2021
Modeling the Evolutionary Architectures of Transcribed Human Enhancer Sequences Reveals Distinct Origins, Functions, and Associations with Human Trait Variation
Fong SL, Capra JA*
Molecular Biology and Evolution,
2021
Evaluating human autosomal loci for sexually antagonistic viability selection in two large biobanks
Kasimatis KR^, Abraham A^, Ralph PL, Kern AD, Capra JA*, Phillips PC.*
Genetics,
2021
Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability
McArthur E, Capra JA.*
Am J Hum Genet,
2021
Genetics of agenesis/hypoplasia of the uterus and vagina: narrowing down the number of candidate genes for Mayer-Rokitansky-Küster-Hauser Syndrome
Mikhael S*, Dugar S, Morton M, Chorich LP, Tam KB, Lossie AC, Kim HG, Knight J, Taylor HS, Mukherjee S, Capra JA, Phillips JA 3rd, Friez M, Layman LC.*
Hum Genet,
2021
The influence of evolutionary history on human health and disease
Benton ML, Abraham A, LaBella AL, Abbot P, Rokas A, Capra JA.*
Nat Rev Genet,
2021
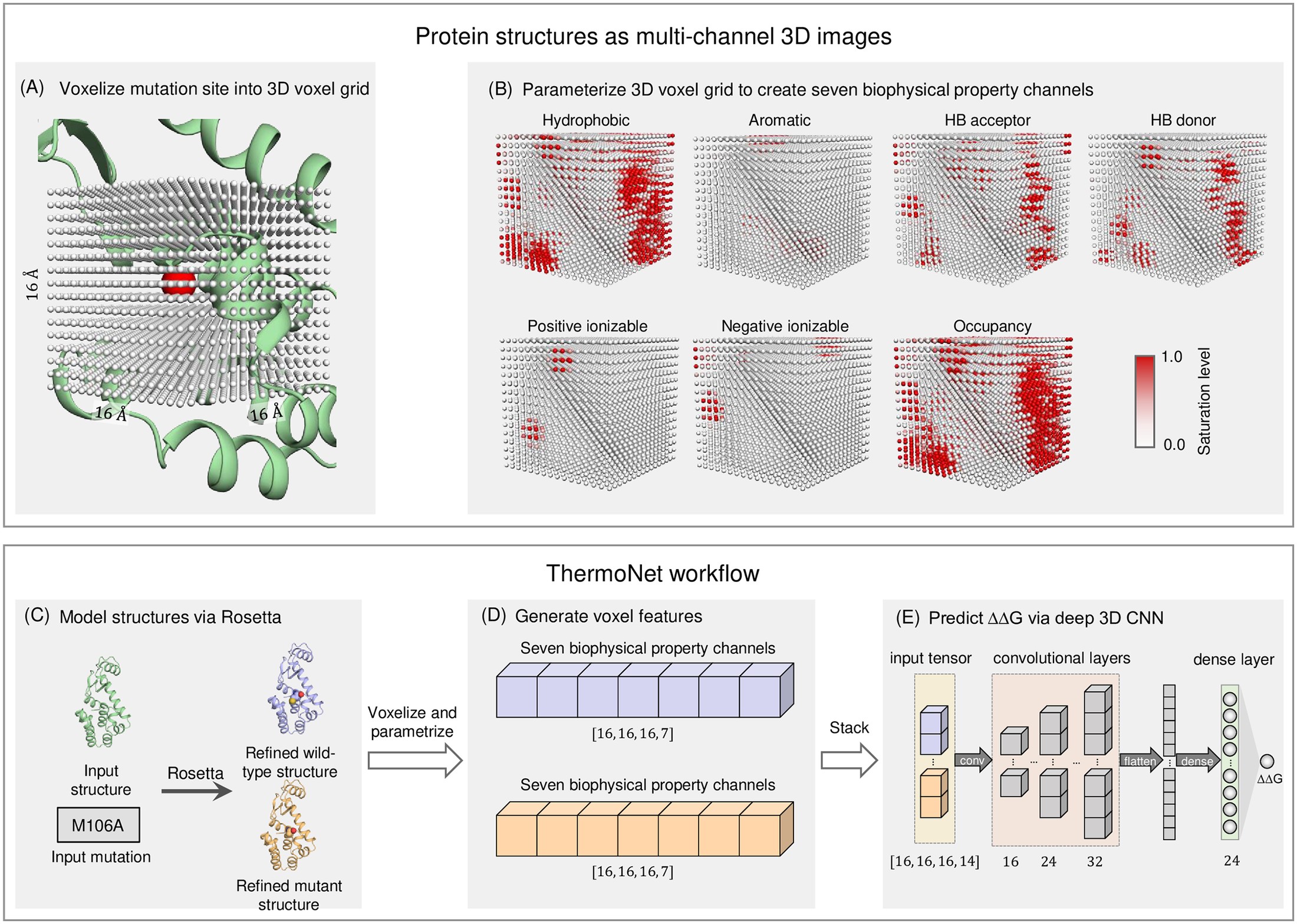
Predicting changes in protein thermodynamic stability upon point mutation with deep 3D convolutional neural networks
Li B, Yang YT, Capra JA*, Gerstein MB.*
PLoS Comput Biol,
2020
Learning and interpreting the gene regulatory grammar in a deep learning framework
Chen L, Capra JA.*
PLoS Comput Biol,
2020
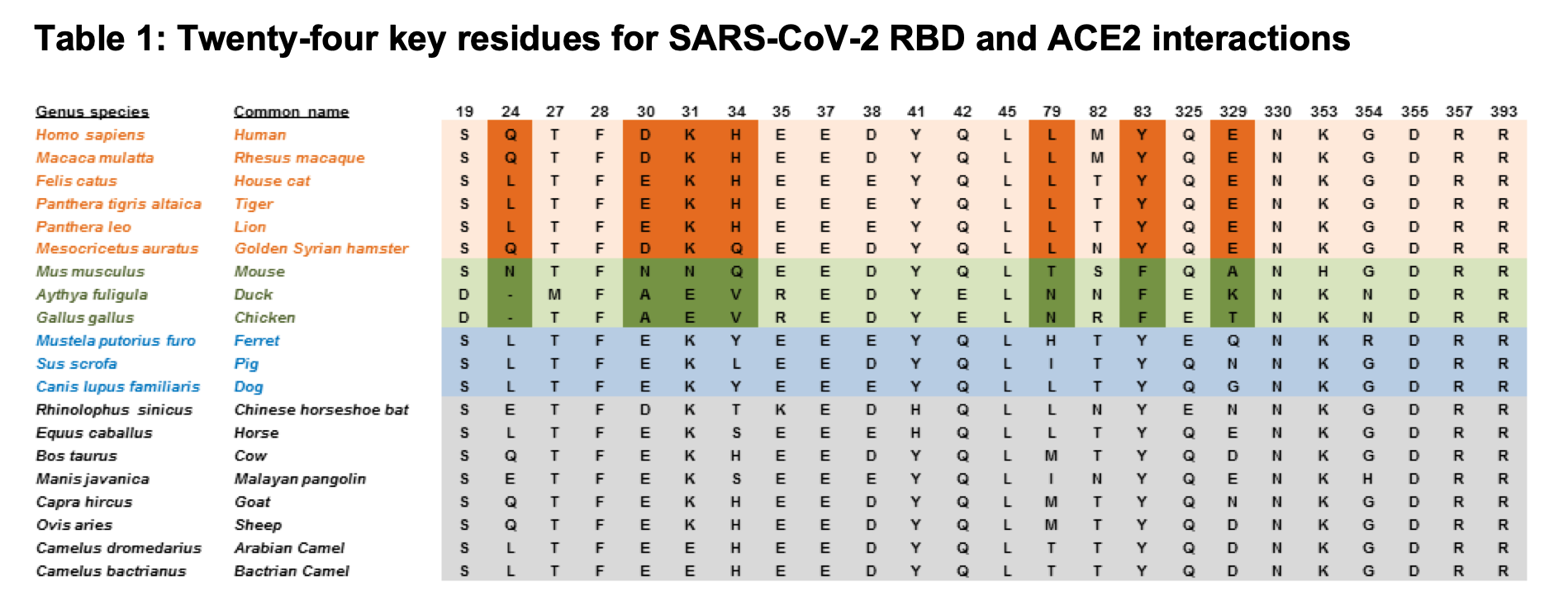
Predicting susceptibility to SARS-CoV-2 infection based on structural differences in ACE2 across species
Alexander MR^, Schoeder CT^, Brown JA, Smart CD, Moth C, Wikswo JP, Capra JA, Meiler J#, Chen W#*, Madhur MS.#*
FASEB J,
2020
PSCAN: Spatial scan tests guided by protein structures improve complex disease gene discovery and signal variant detection
Tang ZZ*, Sliwoski GR, Chen G, Jin B, Bush WS, Li B*, Capra JA.*
Genome Biol,
2020
Phenotypic Profiling in Subjects Heterozygous for 1 of 2 Rare Variants in the Hypophosphatasia Gene (ALPL)
Tilden DR, Sheehan JH, Newman JH, Meiler J, Capra JA, Ramirez A, Simmons J, Dahir K.*
J Endocr Soc,
2020
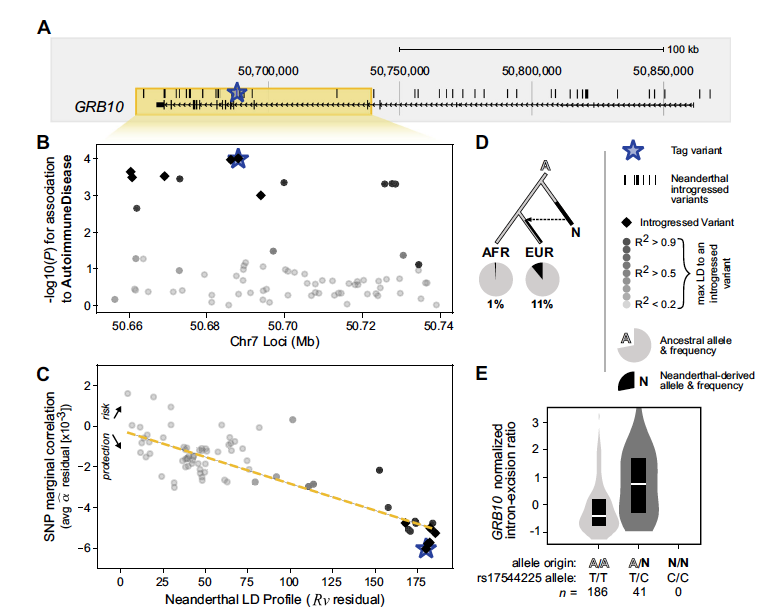
Neanderthal introgression reintroduced functional ancestral alleles lost in Eurasian populations
Rinker DC, Simonti CN, McArthur E, Shaw D, Hodges E, Capra JA.*
Nat Ecol Evol,
2020
Accounting for diverse evolutionary forces reveals mosaic patterns of selection on human preterm birth loci
LaBella AL^, Abraham A^, Pichkar Y, Fong SL, Zhang G, Muglia LJ, Abbot P, Rokas A*, Capra JA.*
Nat Commun,
2020
Which animals are at risk? Predicting species susceptibility to Covid-19
Alexander MR^, Schoeder CT^, Brown JA, Smart CD, Moth C, Wikswo JP, Capra JA, Meiler J*, Chen W*, Madhur MS.*
bioRxiv,
2020
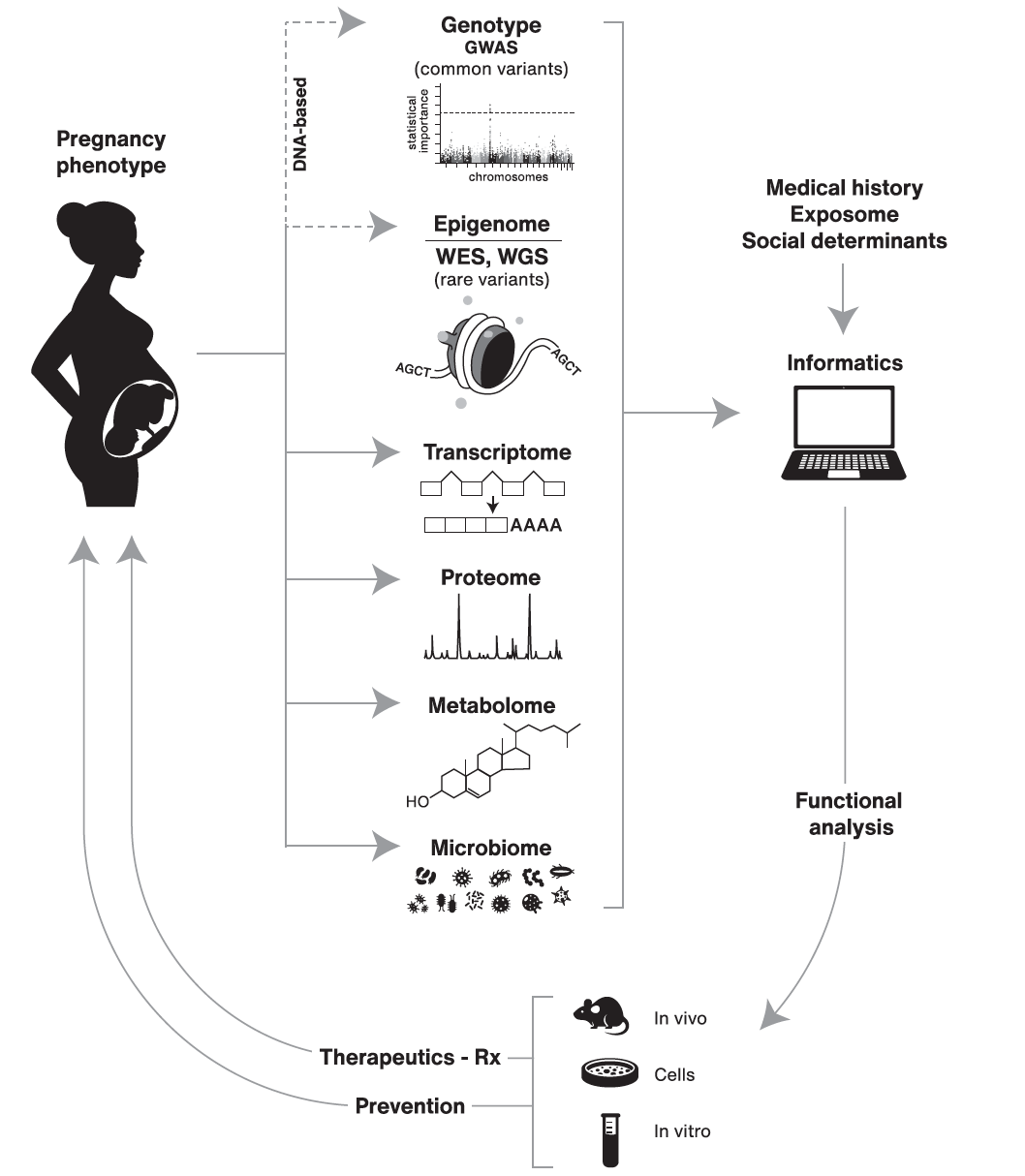
Advancing human health in the decade ahead: pregnancy as a key window for discovery: A Burroughs Wellcome Fund Pregnancy Think Tank
Sadovsky Y, Mesiano S, Burton GJ, Lampl M, Murray JC, Freathy RM, Mahadevan-Jansen A, Moffett A, Price ND, Wise PH, Wildman DE, Snyderman R, Paneth N, Capra JA, Nobrega MA, Barak Y, Muglia LJ*; Burroughs Wellcome Fund Pregnancy Think Tank Working Group.
Am J Obstet Gynecol,
2020
Functional annotation of rare structural variation in the human brain
Han L^, Zhao X^, Benton ML, Perumal T, Collins RL, Hoffman GE, Johnson JS, Sloofman L, Wang HZ, Stone MR; CommonMind Consortium; Brennand KJ, Brand H, Sieberts SK, Marenco S, Peters MA, Lipska BK, Roussos P, Capra JA, Talkowski M, Ruderfer DM.*
Nat Commun,
2020
High-Throughput Reclassification of SCN5A Variants
Glazer AM, Wada Y, Li B, Muhammad A, Kalash OR, O’Neill MJ, Shields T, Hall L, Short L, Blair MA, Kroncke BM, Capra JA, Roden DM.*
Am J Hum Genet,
2020
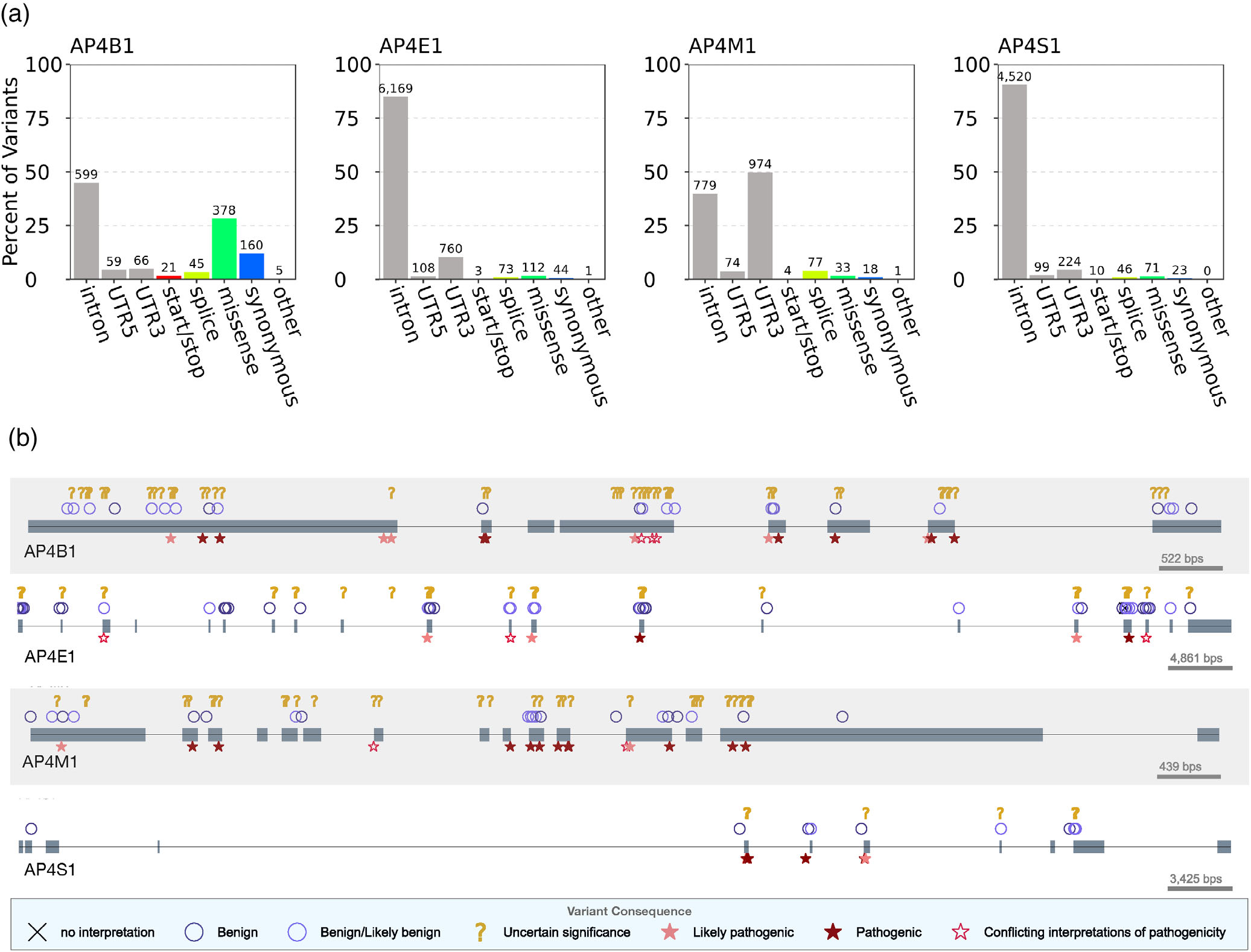
Integrating structural and evolutionary data to interpret variation and pathogenicity in adapter protein complex 4
Gadbery JE^, Abraham A^, Needle CD, Moth C, Sheehan J, Capra JA, Jackson LP.*
Protein Sci,
2020
Inferred divergent gene regulation in archaic hominins reveals potential phenotypic differences
Colbran LL, Gamazon ER, Zhou D, Evans P, Cox NJ, Capra JA.*
Nat Ecol Evol,
2019
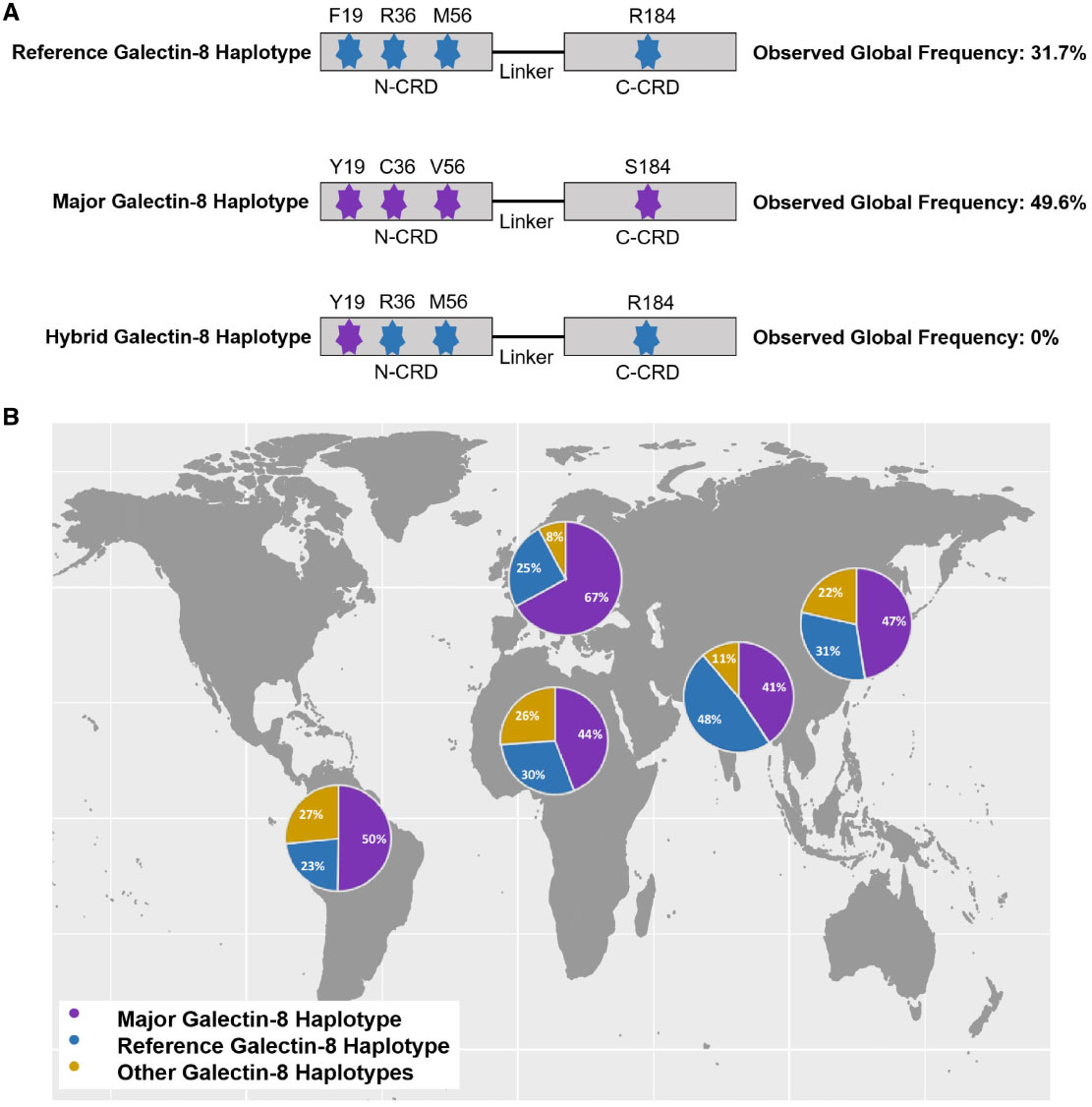
The Impact of Natural Selection on the Evolution and Function of Placentally Expressed Galectins
Ely ZA, Moon JM, Sliwoski GR, Sangha AK, Shen XX, Labella AL, Meiler J, Capra JA, Rokas A.*
Genome Biol Evol,
2019
Immune Regulation in Eutherian Pregnancy: Live Birth Coevolved with Novel Immune Genes and Gene Regulation
Moon JM, Capra JA, Abbot P, Rokas A.*
Bioessays,
2019
Genome-wide association analysis uncovers variants for reproductive variation across dog breeds and links to domestication
Smith SP^, Phillips JB^, Johnson ML, Abbot P, Capra JA, Rokas A.*
Evol Med Public Health,
2019
Genome-wide enhancer annotations differ significantly in genomic distribution, evolution, and function
Benton ML, Talipineni SC, Kostka D*, Capra JA.*
BMC Genomics,
2019
Signatures of Recent Positive Selection in Enhancers Across 41 Human Tissues
Moon JM, Capra JA, Abbot P, Rokas A.*
G3 (Bethesda),
2019
IgG4-related disease: Association with a rare gene variant expressed in cytotoxic T cells
Newman JH*, Shaver A, Sheehan JH, Mallal S, Stone JH, Pillai S, Bastarache L, Riebau D, Allard-Chamard H, Stone WM, Perugino C, Pilkinton M, Smith SA, McDonnell WJ, Capra JA, Meiler J, Cogan J, Xing K, Mahajan VS, Mattoo H, Hamid R, Phillips JA 3rd; Undiagnosed Disease Network.
Mol Genet Genomic Med,
2019
Protein structure aids predicting functional perturbation of missense variants in SCN5A and KCNQ1
Kroncke BM*, Mendenhall J, Smith DK, Sanders CR, Capra JA, George AL, Blume JD, Meiler J, Roden DM.
Comput Struct Biotechnol J,
2019
Sequence Characteristics Distinguish Transcribed Enhancers from Promoters and Predict Their Breadth of Activity
Colbran LL, Chen L, Capra JA.*
Genetics,
2019
Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis
Marinko JT^, Huang H^, Penn WD, Capra JA, Schlebach JP*, Sanders CR.*
Chem Rev,
2019
Genome-wide maps of distal gene regulatory enhancers active in the human placenta
Zhang J^, Simonti CN^, Capra JA.*
PLoS One,
2018
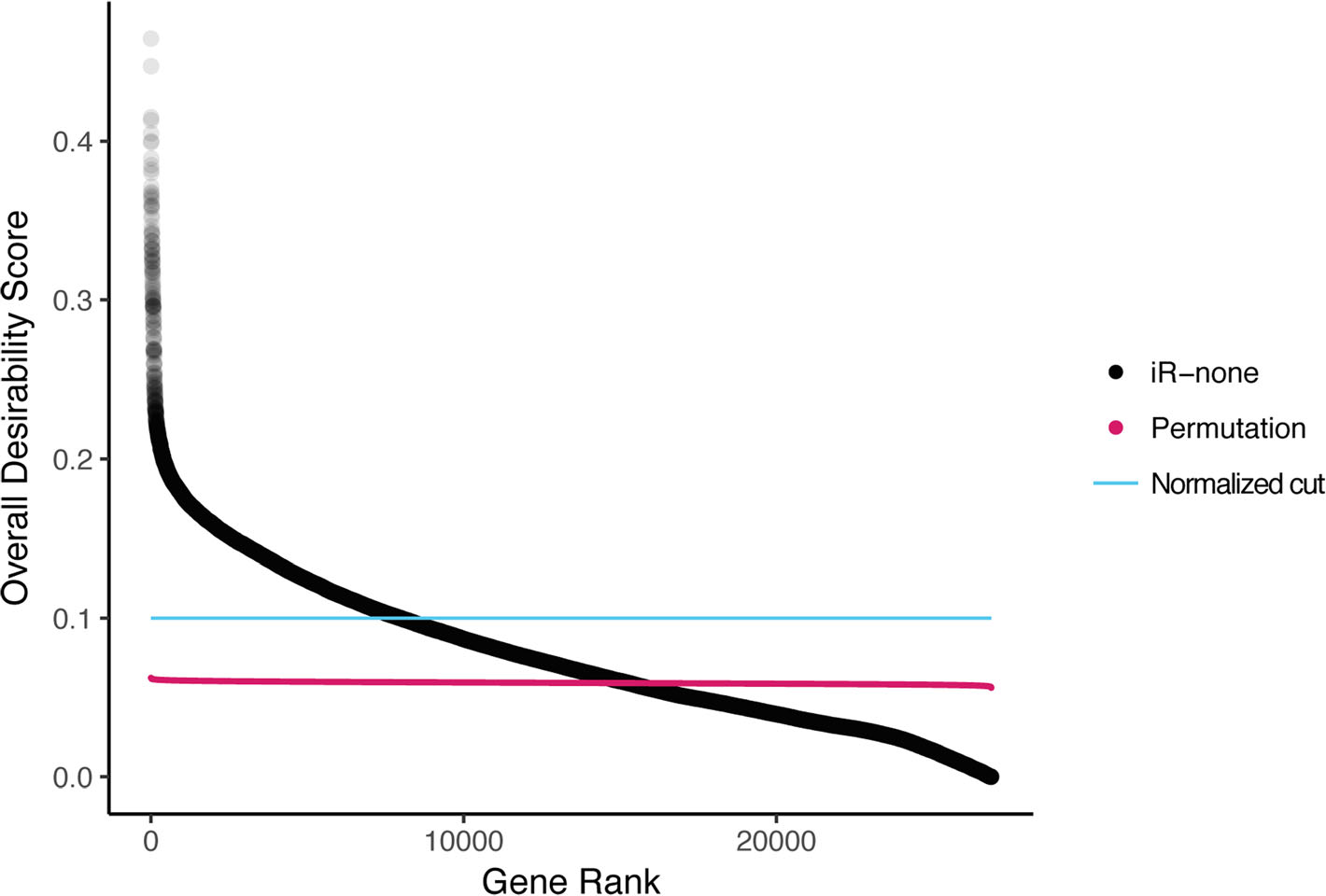
integRATE: a desirability-based data integration framework for the prioritization of candidate genes across heterogeneous omics and its application to preterm birth
Eidem HR, Steenwyk JL, Wisecaver JH, Capra JA, Abbot P, Rokas A.*
BMC Med Genomics,
2018
Prediction of gene regulatory enhancers across species reveals evolutionarily conserved sequence properties
Chen L^, Fish AE^, Capra JA.*
PLoS Comput Biol,
2018
Examination of Signatures of Recent Positive Selection on Genes Involved in Human Sialic Acid Biology
Moon JM, Aronoff DM, Capra JA, Abbot P*, Rokas A.*
G3 (Bethesda),
2018
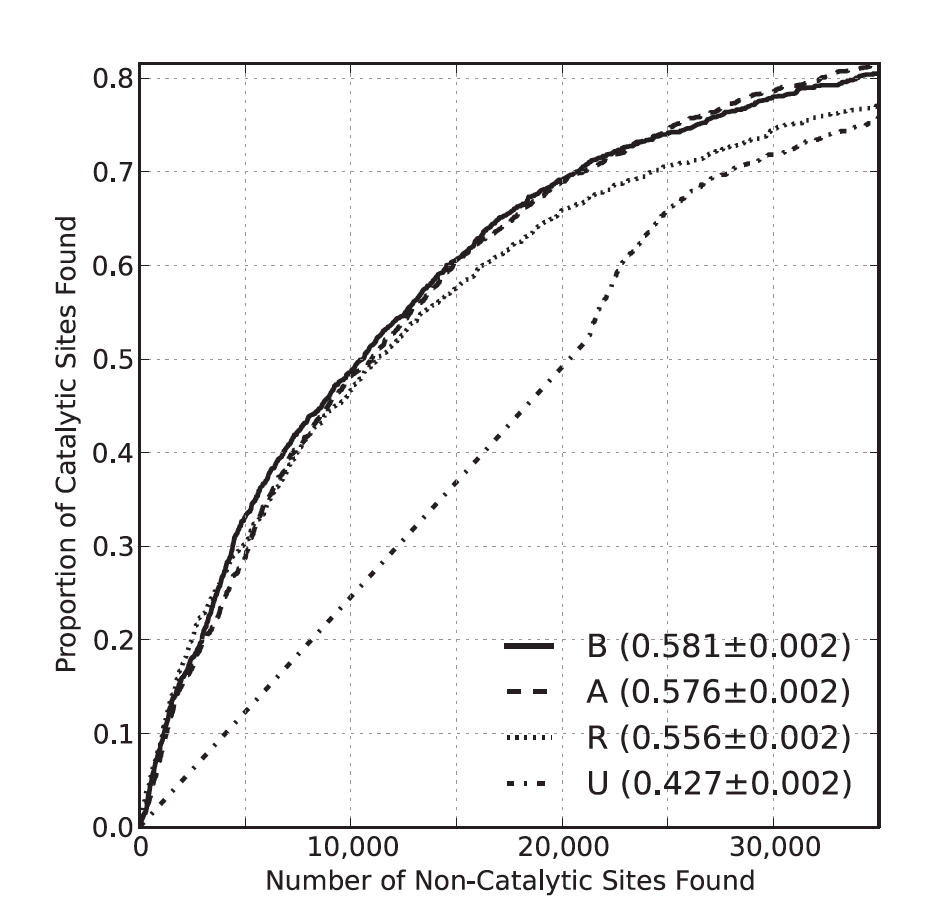
Comprehensive Analysis of Constraint on the Spatial Distribution of Missense Variants in Human Protein Structures
Sivley RM, Dou X, Meiler J, Bush WS*, Capra JA.*
Am J Hum Genet,
2018
Three-dimensional spatial analysis of missense variants in RTEL1 identifies pathogenic variants in patients with Familial Interstitial Pneumonia
Sivley RM, Sheehan JH, Kropski JA, Cogan J, Blackwell TS, Phillips JA, Bush WS, Meiler J, Capra JA.*
BMC Bioinformatics,
2018
Local ancestry transitions modify snp-trait associations
Fish AE, Crawford DC, Capra JA^, Bush WS.^
Pac Symp Biocomput,
2017
Gene Regulatory Enhancers with Evolutionarily Conserved Activity Are More Pleiotropic than Those with Species-Specific Activity
Fish A^, Chen L^, Capra JA.*
Genome Biol Evol,
2017
Transposable Element Exaptation into Regulatory Regions Is Rare, Influenced by Evolutionary Age, and Subject to Pleiotropic Constraints
Simonti CN, Pavlicev M, Capra JA.*
Mol Biol Evol,
2017
What is a placental mammal anyway?
Abbot P^*, Capra JA.^*
Elife,
2017
The transformative potential of an integrative approach to pregnancy
Eidem HR^, McGary KL^, Capra JA, Abbot P, Rokas A.*
Placenta,
2017
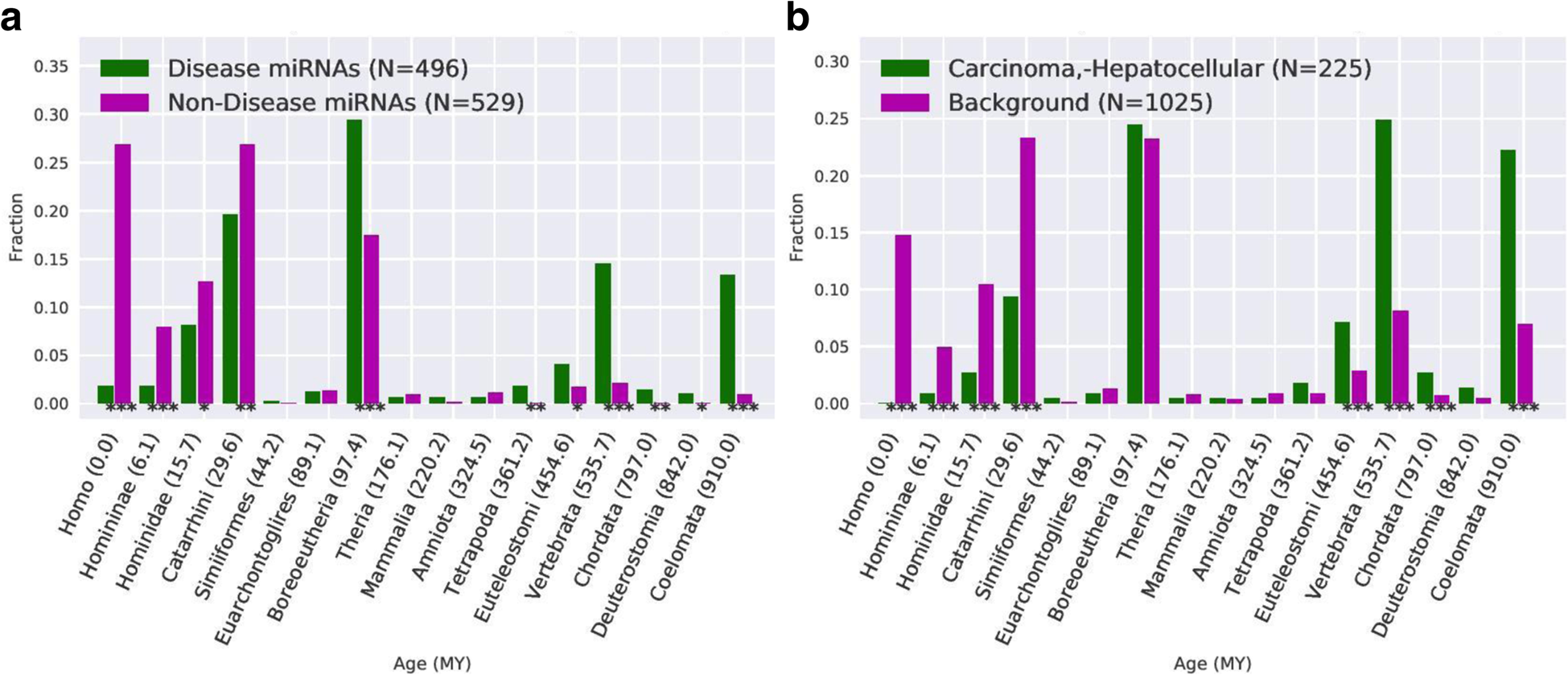
Ancient human miRNAs are more likely to have broad functions and disease associations than young miRNAs
Patel VD, Capra JA.*
BMC Genomics,
2017
Short DNA sequence patterns accurately identify broadly active human enhancers
Colbran LL, Chen L, Capra JA.*
BMC Genomics,
2017
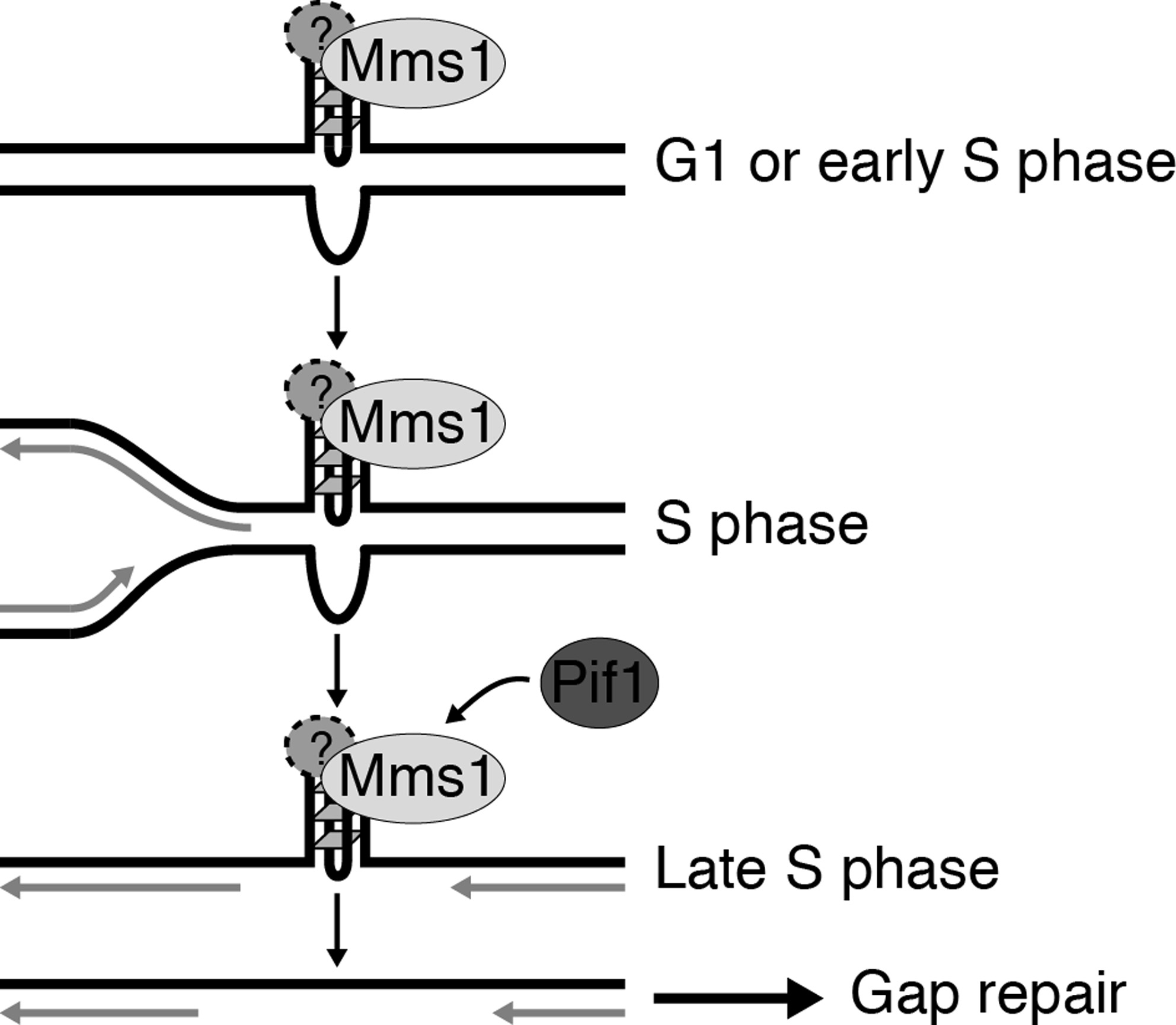
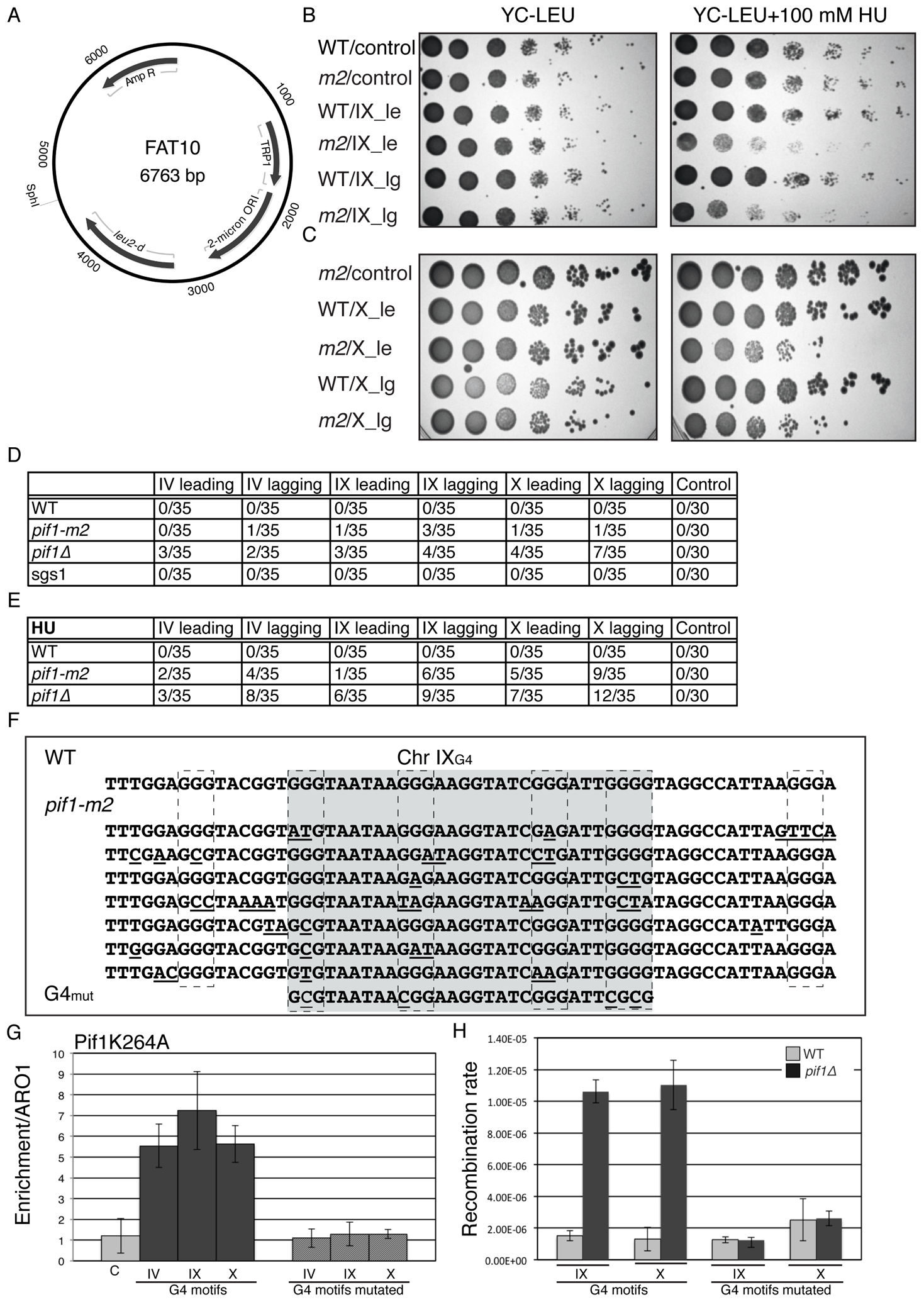
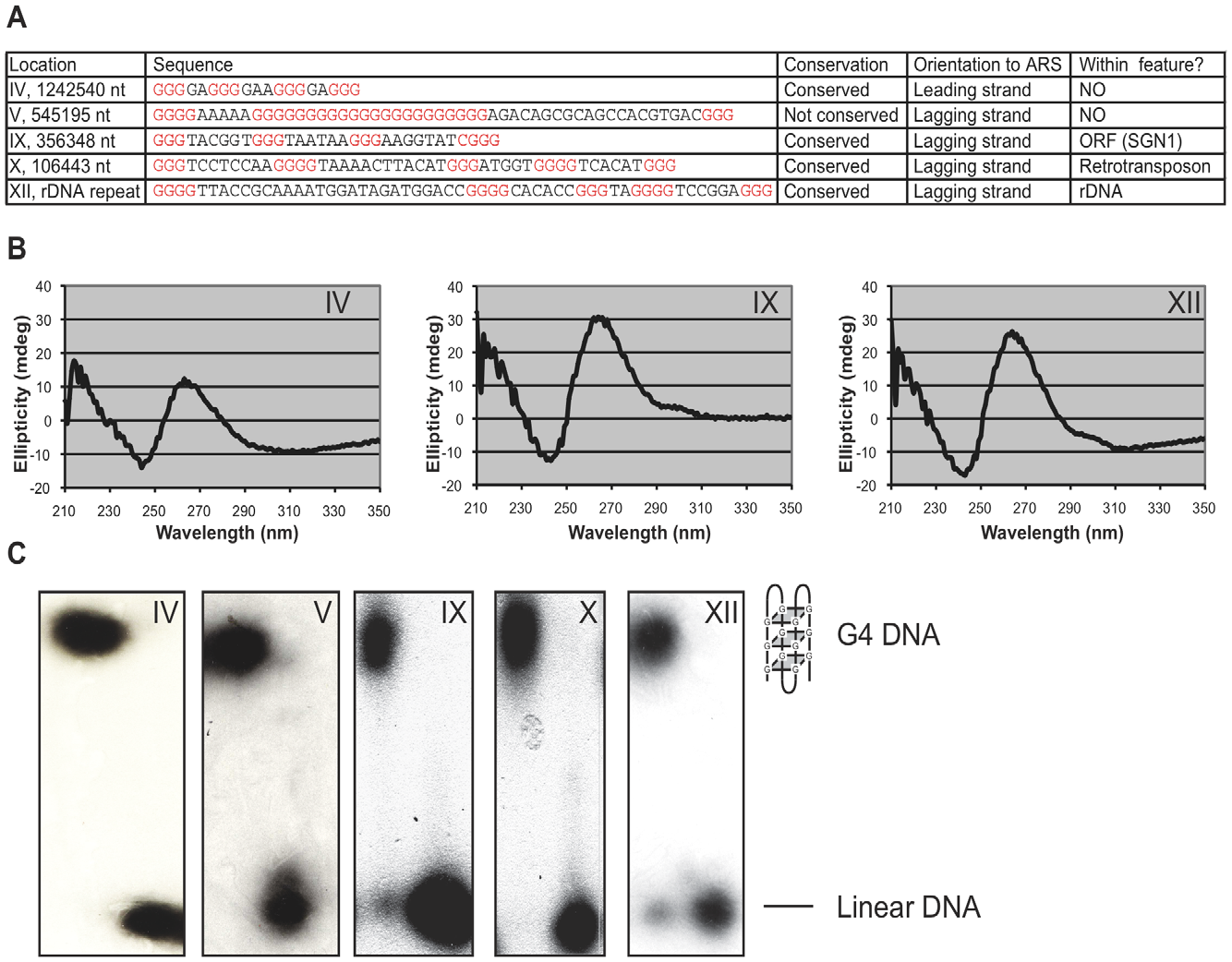
Mms1 binds to G-rich regions in Saccharomyces cerevisiae and influences replication and genome stability
Wanzek K, Schwindt E, Capra JA, Paeschke K.*
Nucleic Acids Res,
2017
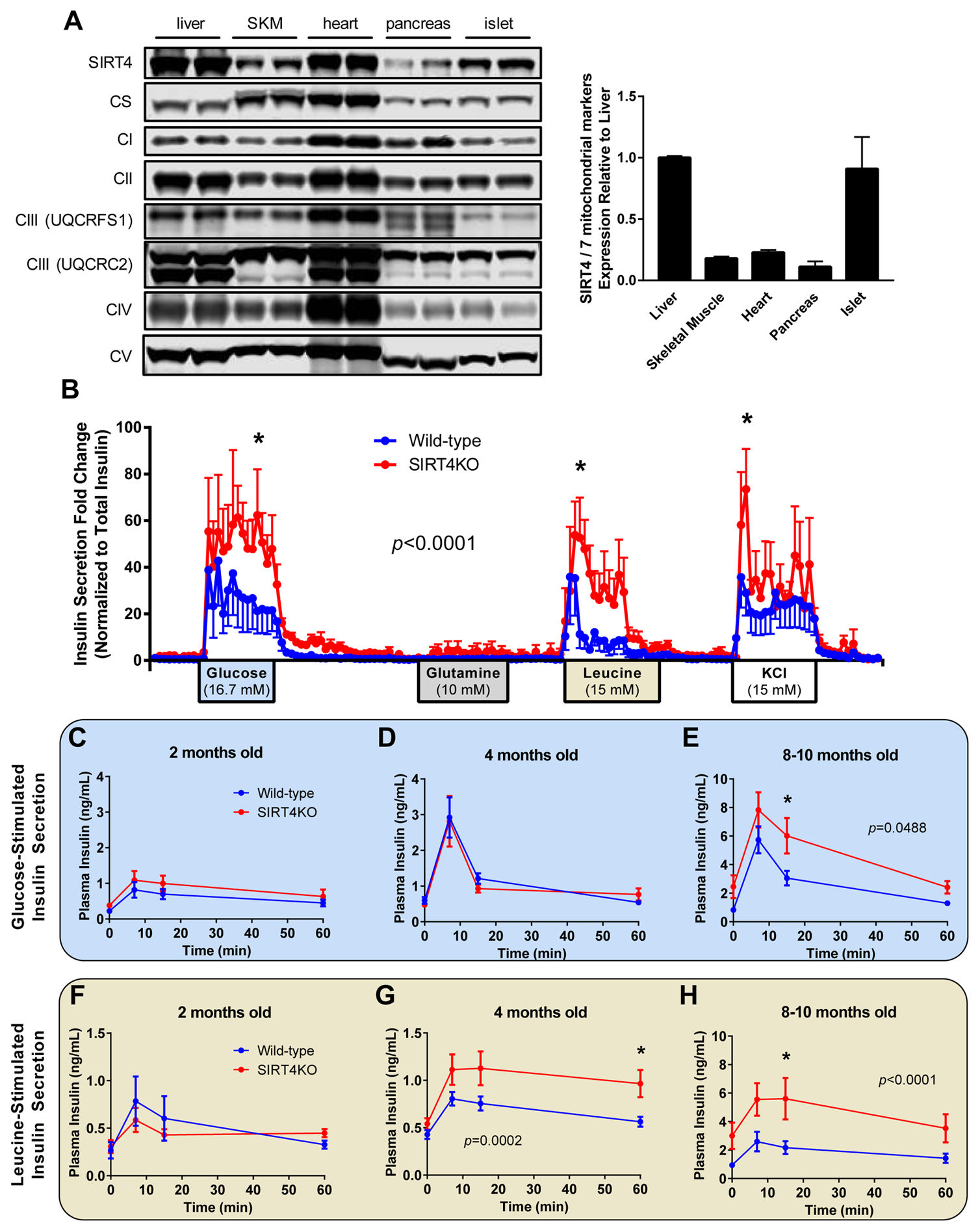
SIRT4 Is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion
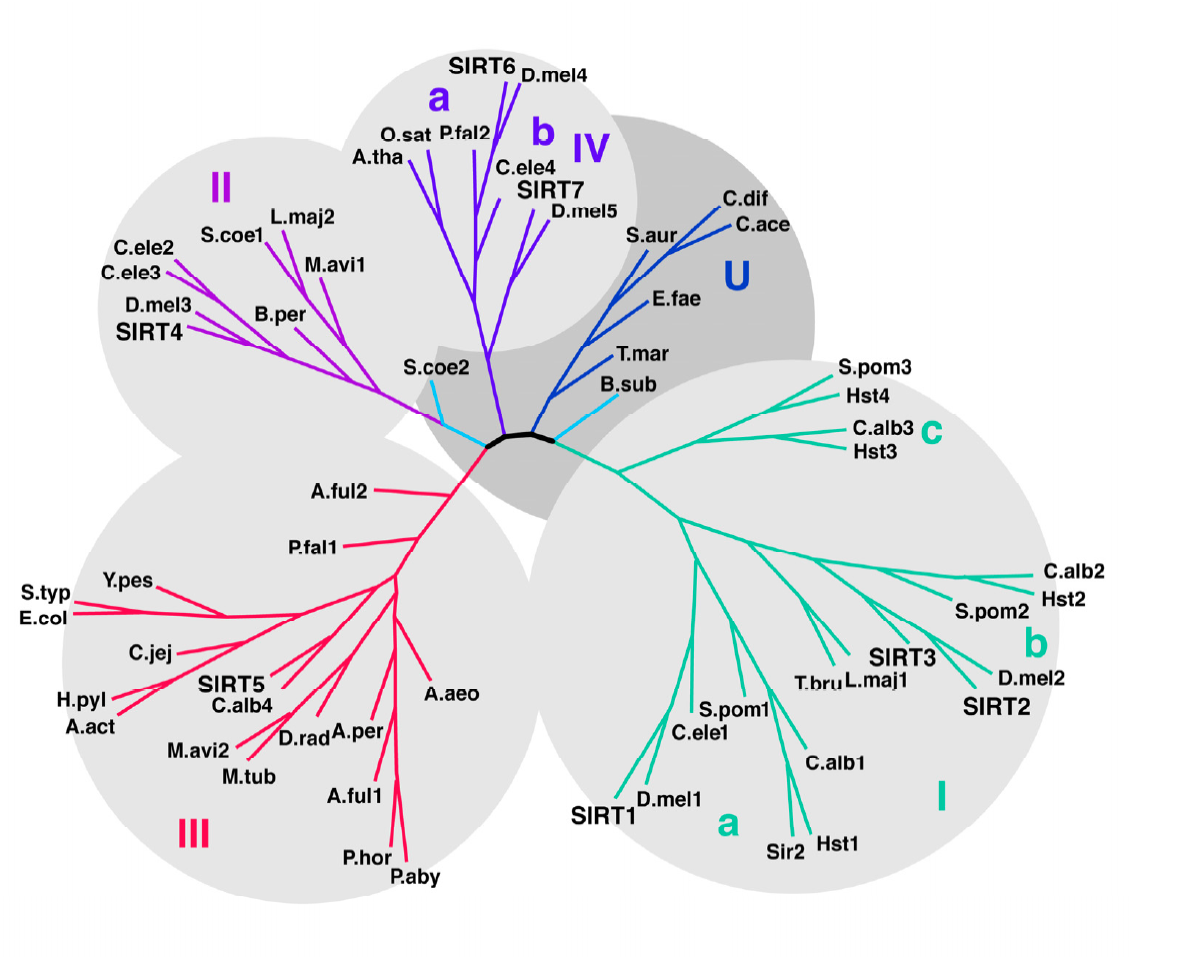
Anderson KA^, Huynh FK^, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, Wagner GR, Thompson JW, Madsen AS, Green MF, Sivley RM, Ilkayeva OR, Stevens RD, Backos DS, Capra JA, Olsen CA, Campbell JE, Muoio DM, Grimsrud PA, Hirschey MD.*
Cell Metab,
2017
Are Interactions between cis-Regulatory Variants Evidence for Biological Epistasis or Statistical Artifacts?
Fish AE, Capra JA, Bush WS.*
Am J Hum Genet,
2016
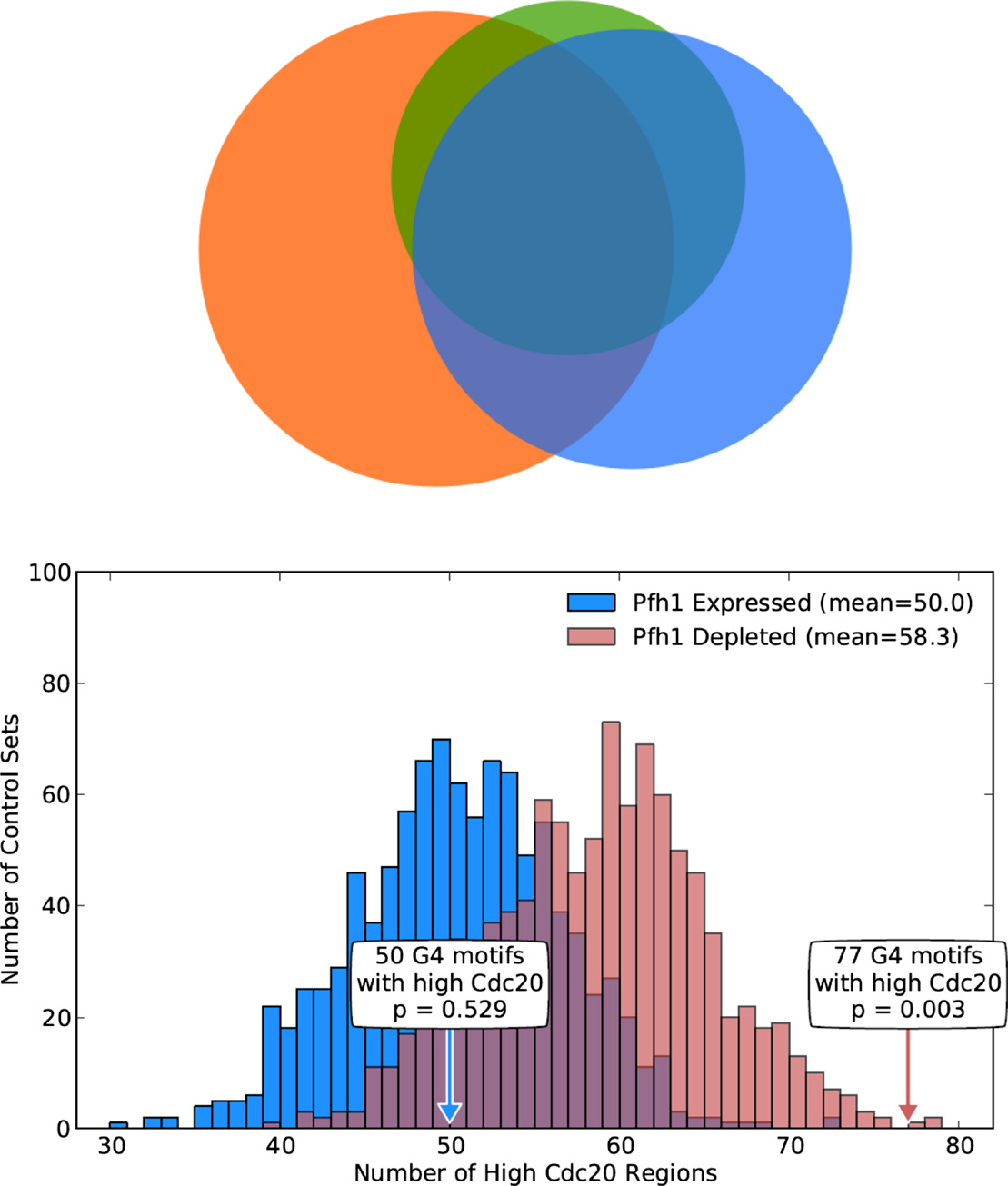
Pfh1 Is an Accessory Replicative Helicase that Interacts with the Replisome to Facilitate Fork Progression and Preserve Genome Integrity
McDonald KR, Guise AJ, Pourbozorgi-Langroudi P, Cristea IM, Zakian VA, Capra JA*, Sabouri N.*
PLoS Genet,
2016
Heterozygosity Ratio, a Robust Global Genomic Measure of Autozygosity and Its Association with Height and Disease Risk
Samuels DC^, Wang J^, Ye F, He J, Levinson RT, Sheng Q, Zhao S, Capra JA, Shyr Y, Zheng W, Guo Y.*
Genetics,
2016
XPA: A key scaffold for human nucleotide excision repair
Sugitani N, Sivley RM, Perry KE, Capra JA, Chazin WJ.*
DNA Repair (Amst),
2016
The phenotypic legacy of admixture between modern humans and Neandertals
Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, Crosslin DR, Hebbring SJ, Jarvik GP, Kullo IJ, Li R, Pathak J, Ritchie MD, Roden DM, Verma SS, Tromp G, Prato JD, Bush WS, Akey JM^, Denny JC^, Capra JA*
Science,
2016
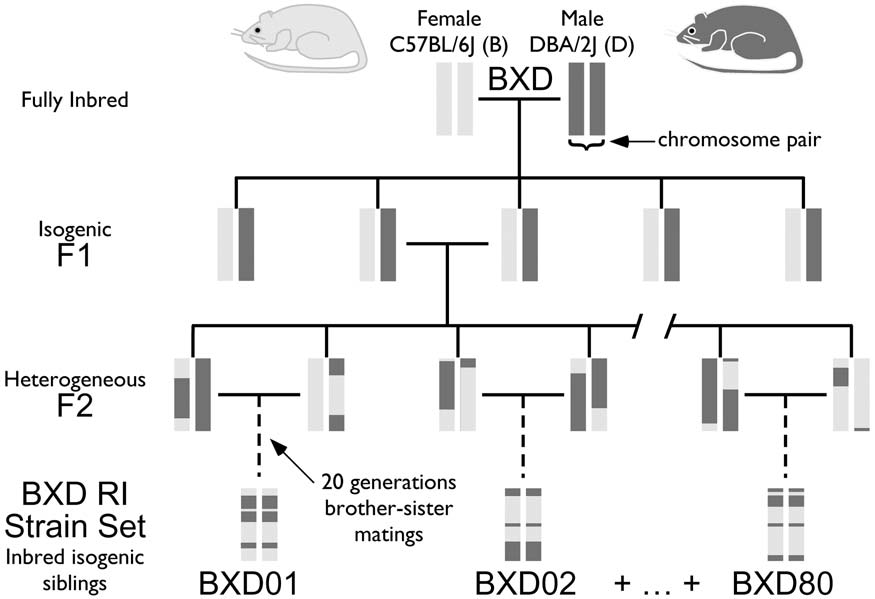
Joint mouse-human phenome-wide association to test gene function and disease risk
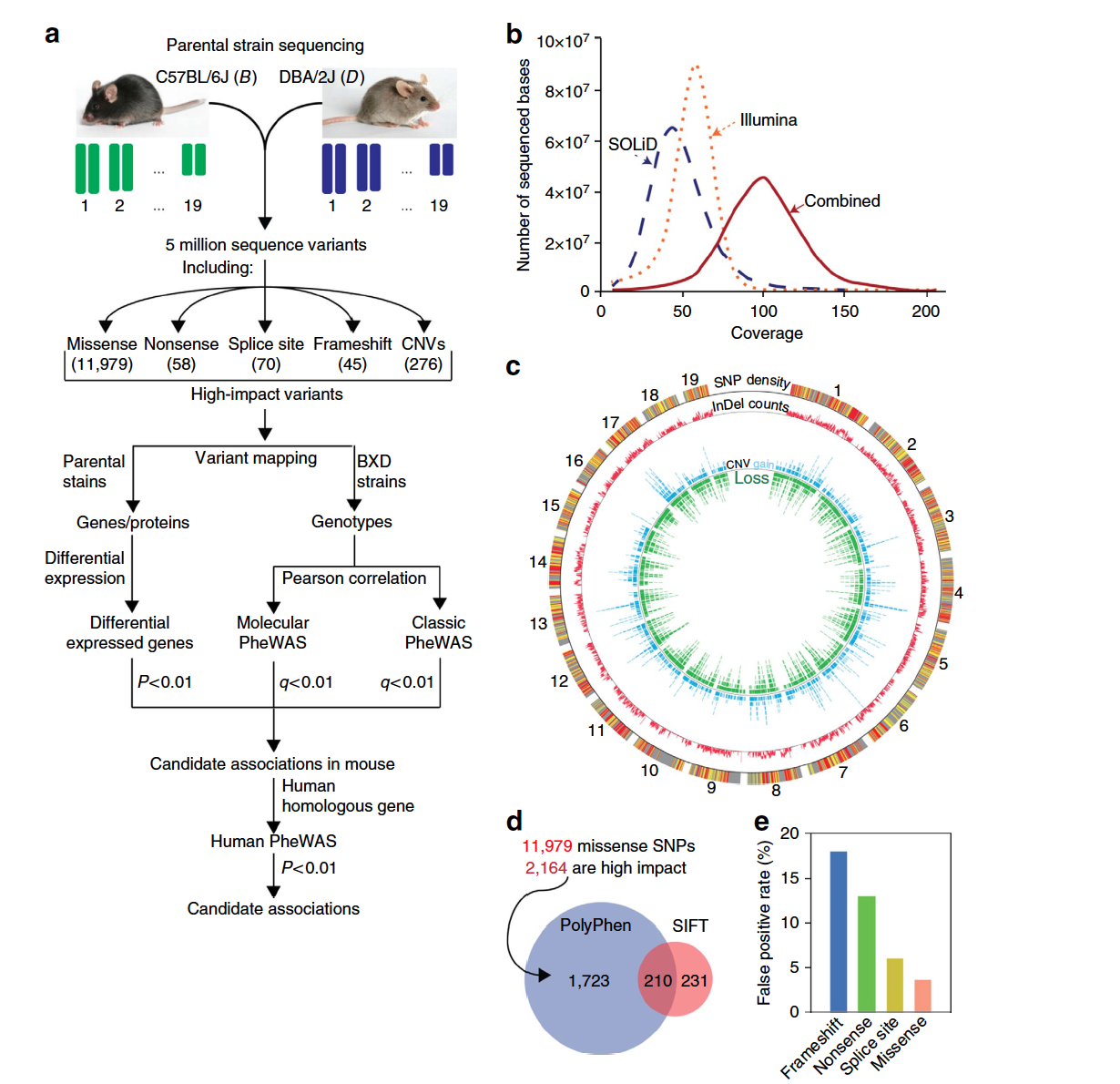
Wang X^, Pandey AK^, Mulligan MK, Williams EG, Mozhui K, Li Z, Jovaisaite V, Quarles LD, Xiao Z, Huang J, Capra JA, Chen Z, Taylor WL, Bastarache L, Niu X, Pollard KS, Ciobanu DC, Reznik AO, Tishkov AV, Zhulin IB, Peng J, Nelson SF, Denny JC, Auwerx J, Lu L, Williams RW.*
Nat Commun,
2016
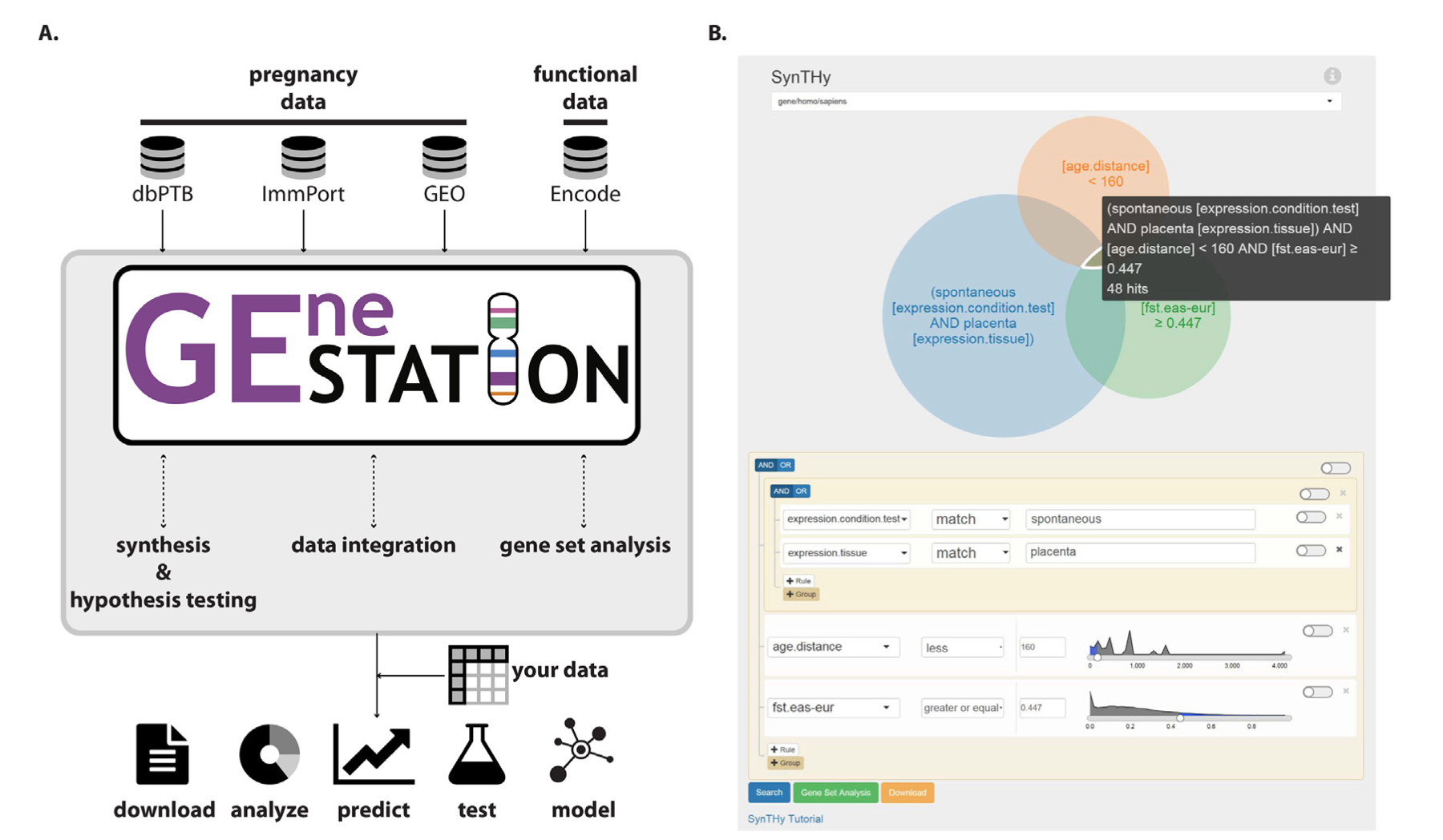
GEneSTATION 1.0: a synthetic resource of diverse evolutionary and functional genomic data for studying the evolution of pregnancy-associated tissues and phenotypes
Kim M, Cooper BA, Venkat R, Phillips JB, Eidem HR, Hirbo J, Nutakki S, Williams SM, Muglia LJ, Capra JA, Petren K, Abbot P, Rokas A*, McGary KL.
Nucleic Acids Res,
2015
The evolution of the human genome
Simonti CN, Capra JA.*
Curr Opin Genet Dev,
2015
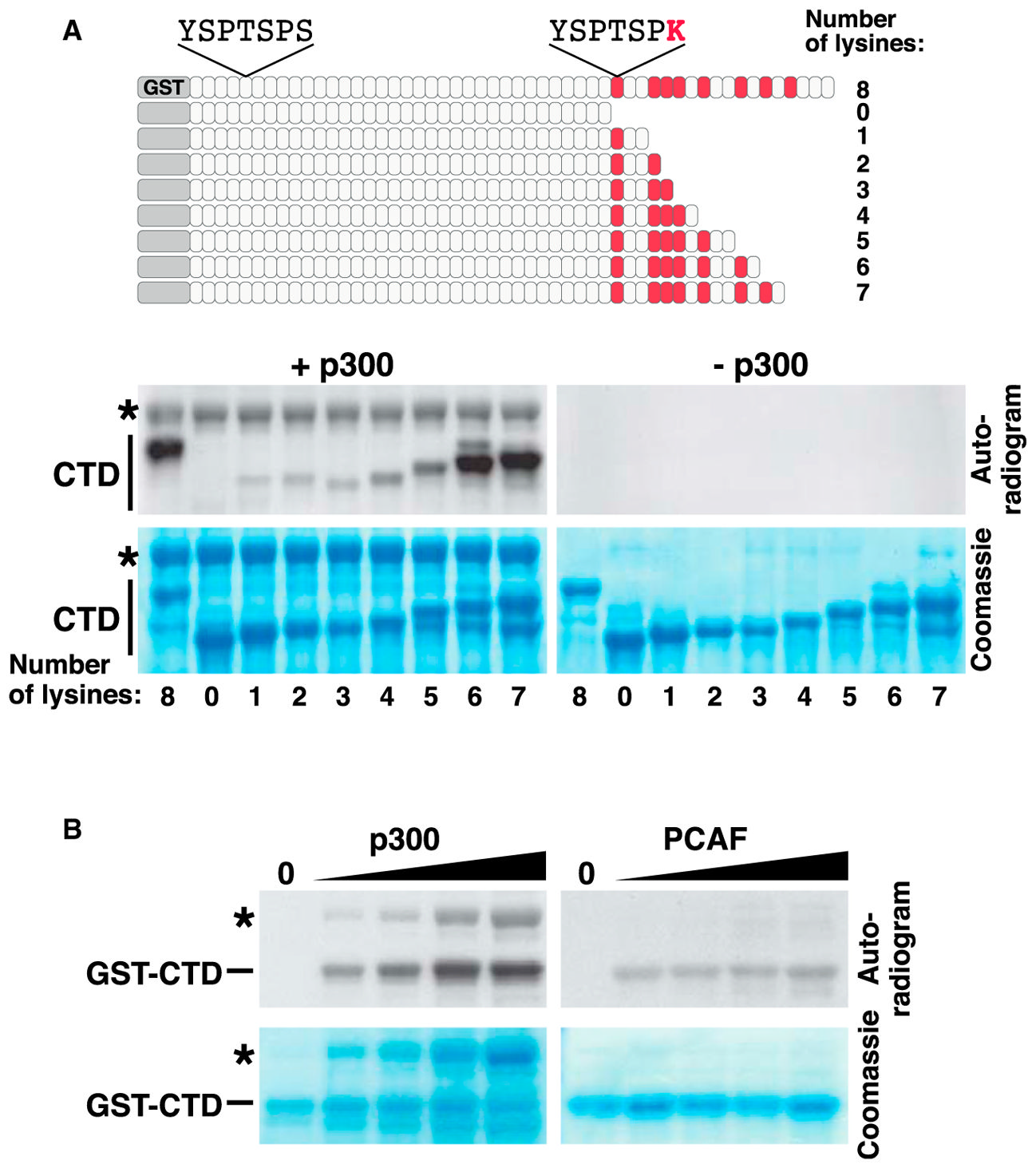
Evolution of lysine acetylation in the RNA polymerase II C-terminal domain
Simonti CN^, Pollard KS^, Schröder S^, He D, Bruneau BG, Ott M, Capra JA.*
BMC Evol Biol,
2015
Extrapolating histone marks across developmental stages, tissues, and species: an enhancer prediction case study
Capra JA.
BMC Genomics,
2015
The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage
Sabouri N^*, Capra JA^*, Zakian VA.
BMC Biol,
2014
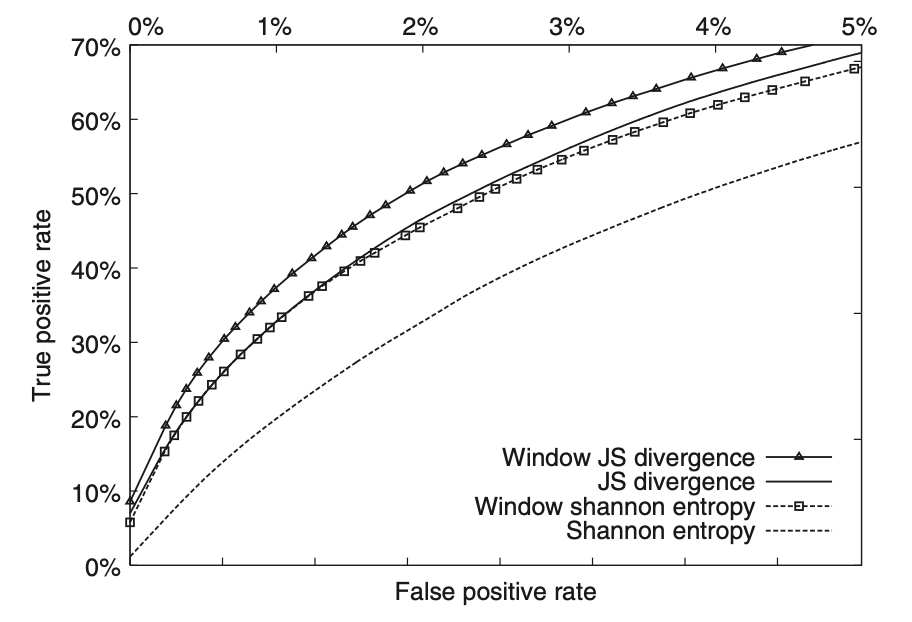
Log-odds sequence logos
Yu YK, Capra JA, Stojmirović A, Landsman D, Altschul SF.*
Bioinformatics,
2014
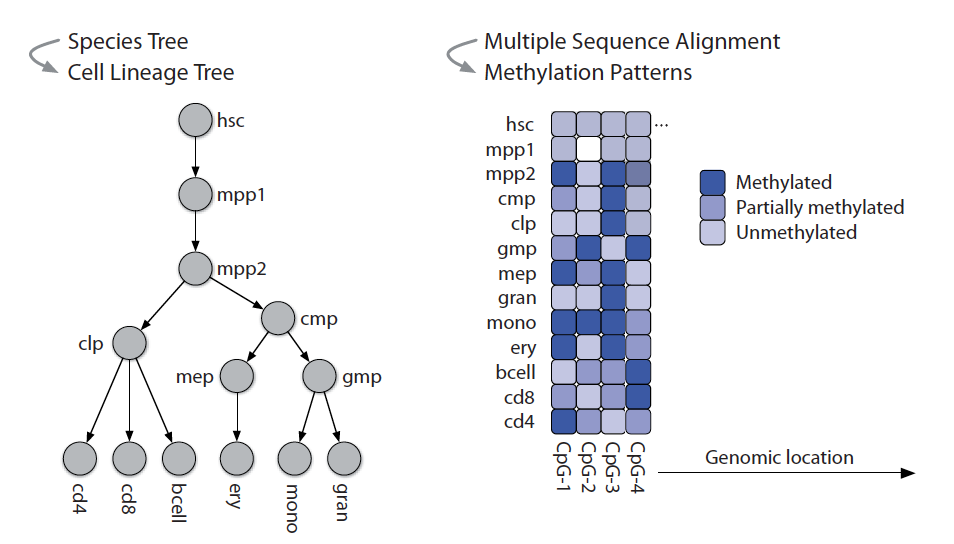
Modeling DNA methylation dynamics with approaches from phylogenetics
Capra JA*, Kostka D.*
Bioinformatics,
2014
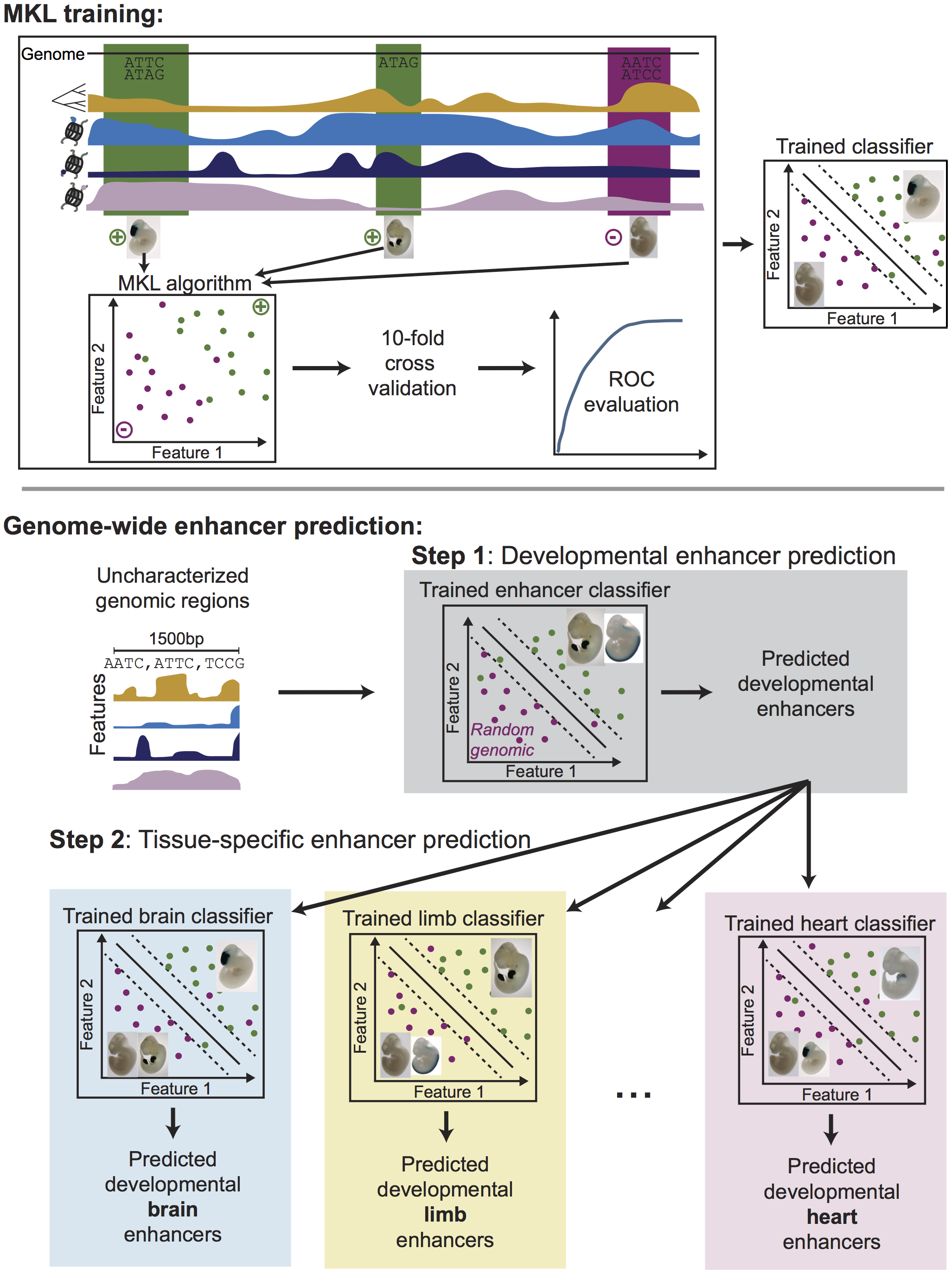
Integrating diverse datasets improves developmental enhancer prediction
Erwin GD, Oksenberg N, Truty RM, Kostka D, Murphy KK, Ahituv N, Pollard KS*, Capra JA.*
PLoS Comput Biol,
2014
Many human accelerated regions are developmental enhancers
Capra JA*, Erwin GD, McKinsey G, Rubenstein JL, Pollard KS.*
Philos Trans R Soc Lond B Biol Sci,
2013
Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells
Schröder S, Herker E, Itzen F, He D, Thomas S, Gilchrist DA, Kaehlcke K, Cho S, Pollard KS, Capra JA, Schnölzer M, Cole PA, Geyer M, Bruneau BG, Adelman K, Ott M.*
Mol Cell,
2013
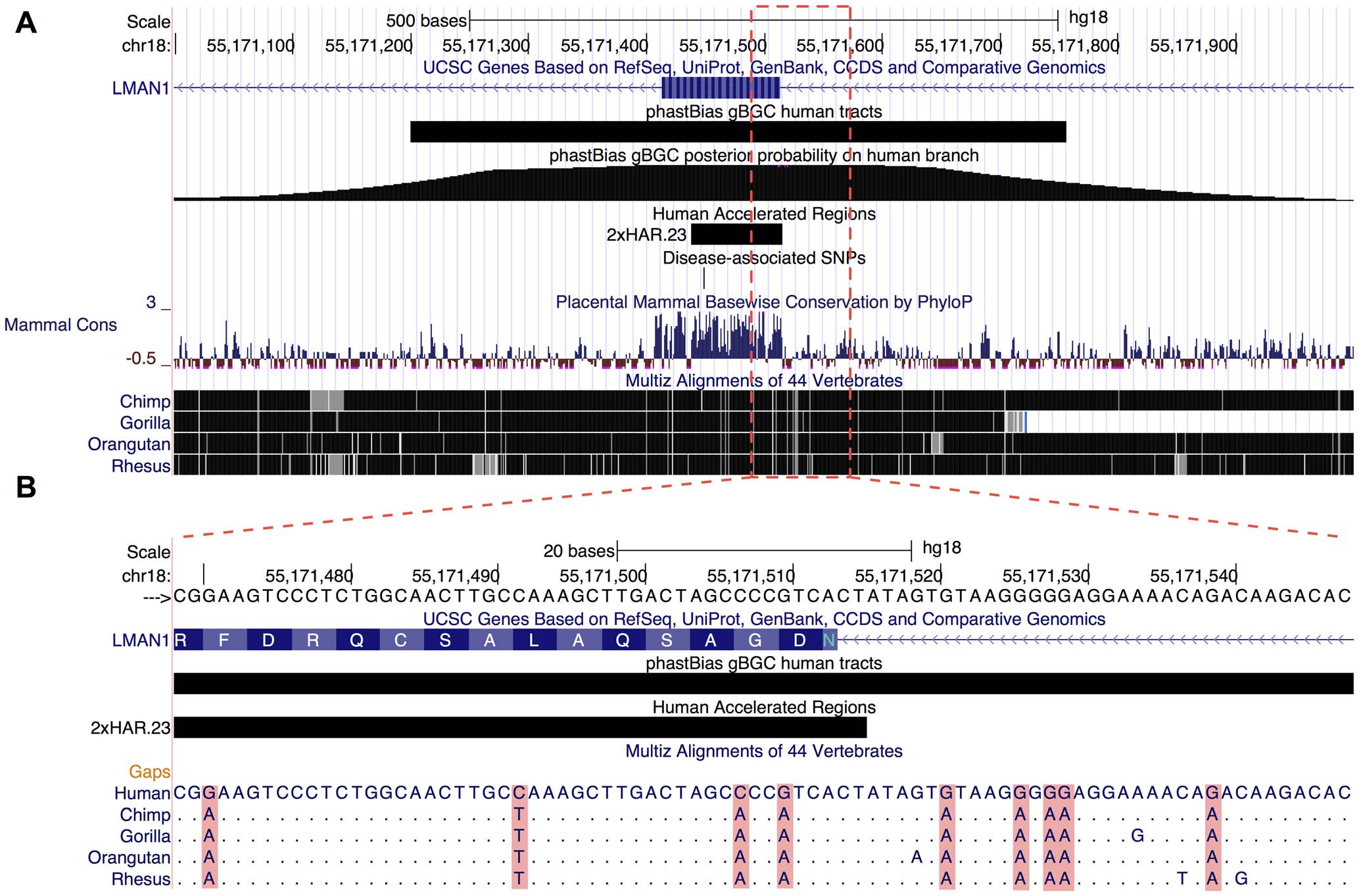
A model-based analysis of GC-biased gene conversion in the human and chimpanzee genomes
Capra JA^, Hubisz MJ^, Kostka D, Pollard KS*, Siepel A.*
PLoS Genet,
2013
How old is my gene?
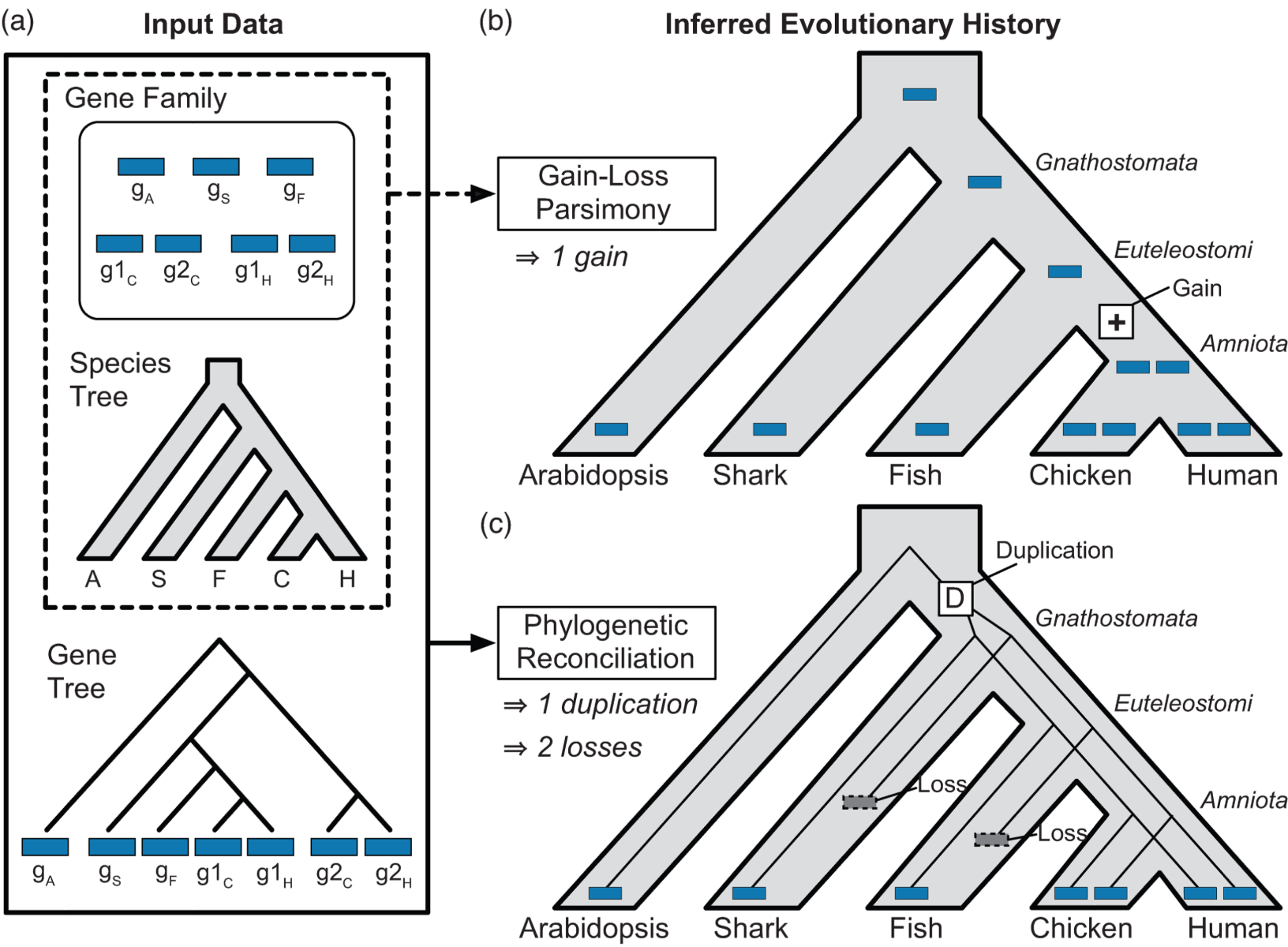
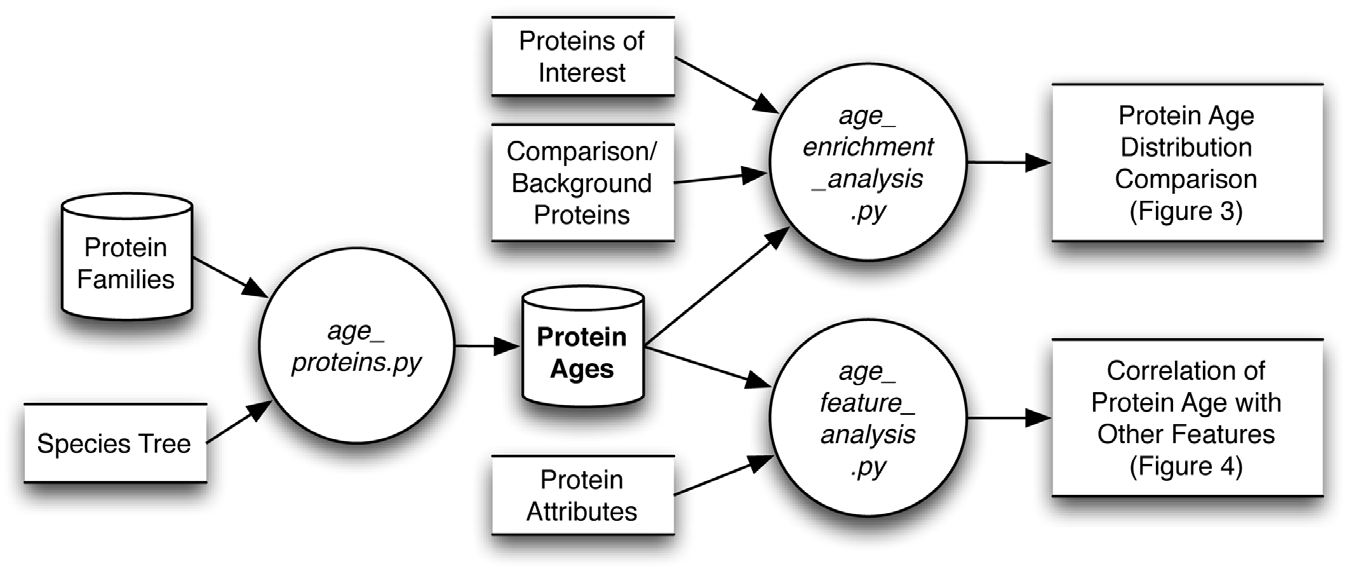
Capra JA, Stolzer M, Durand D*, Pollard KS.*
Trends Genet,
2013
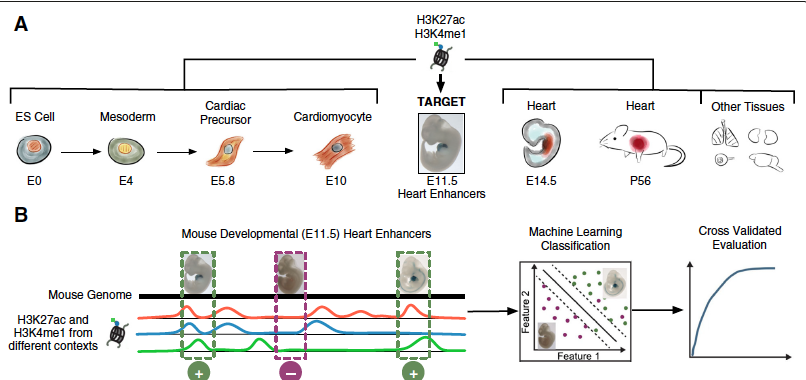
Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage
Wamstad JA^, Alexander JM^, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA*, Bruneau BG.*
Cell,
2012
ProteinHistorian: tools for the comparative analysis of eukaryote protein origin
Capra JA*, Williams AG, Pollard KS.
PLoS Comput Biol,
2012
SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2
Hirschey MD^, Shimazu T^, Capra JA^, Pollard KS, Verdin E.^*
Aging (Albany NY),
2011
Ongoing GC-biased evolution is widespread in the human genome and enriched near recombination hot spots
Katzman S^, Capra JA^, Haussler D, Pollard KS.*
Genome Biol Evol,
2011
Substitution patterns are GC-biased in divergent sequences across the metazoans
Capra JA, Pollard KS.*
Genome Biol Evol,
2011
DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase
Paeschke K, Capra JA, Zakian VA.*
Cell,
2011
Novel genes exhibit distinct patterns of function acquisition and network integration
Capra JA, Pollard KS*, Singh M.*
Genome Biol,
2010
G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae
Capra JA^, Paeschke K^, Singh M*, Zakian VA.*
PLoS Comput Biol,
2010
Genomics through the lens of next-generation sequencing
Capra JA, Carbone L, Riesenfeld SJ, Wall JD.*
Genome Biol,
2010
Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure
Capra JA, Laskowski RA, Thornton JM, Singh M*, Funkhouser TA.*
PLoS Comput Biol,
2009
Characterization and prediction of residues determining protein functional specificity
Capra JA, Singh M.*
Bioinformatics,
2008
Predicting functionally important residues from sequence conservation
Capra JA, Singh M.*
bioinf,
2007/05/24
Informatics center for mouse genomics: the dissection of complex traits of the nervous system
Rosen GD*, La Porte NT, Diechtiareff B, Pung CJ, Nissanov J, Gustafson C, Bertrand L, Gefen S, Fan Y, Tretiak OJ, Manly KF, Park MR, Williams AG, Connolly MT, Capra JA, Williams RW.
Neuroinformatics,
2004